| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,727,699 – 18,727,795 |
| Length | 96 |
| Max. P | 0.978592 |

| Location | 18,727,699 – 18,727,795 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 68.17 |
| Shannon entropy | 0.62465 |
| G+C content | 0.37473 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -10.98 |
| Energy contribution | -10.57 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18727699 96 - 21146708 -ACUGUGCUCAGCACCAG---------CAACCGAAAGAGCCGCCAUAAAUGAAUGAGUUUAUUUA-UUUUUUGGCAUAGUAUUCUUUAUGGCCGCAAUCUGGUCGAA -.(((....))).(((((---------....(....).((.((((((((.(((((.(((......-......)))....))))))))))))).))...))))).... ( -23.30, z-score = -0.91, R) >droSim1.chr2R 17323445 105 - 19596830 -ACUCUGCUCAGCACCAGCACUUGGGCCAACCGAAAGAGCCGCCAUAAAUGAAUGAGUUUAUUUA-UUUUUUGGCAUAGUAUUCUUUAUGGCCGCAAUCUGGUCGAA -..........(.(((((...((((((((..(....)....((((.(((((((((....))))))-)))..)))).............))))).))).))))))... ( -29.40, z-score = -1.62, R) >droSec1.super_9 2028131 103 - 3197100 -ACUCUGCUCAGCACCAGCACC--AGCCAACCGAAAGAGCCGCCAUAAAUGAAUGAGUGUAUUUA-UUUUUUGGUAUAGUAUUCUUUAUGGCCGCAAUCUAGUCGAA -.....(((..((....))...--)))....(((.(((((.((((((((.(((((..((((((..-......)))))).))))))))))))).))..)))..))).. ( -25.10, z-score = -1.68, R) >droYak2.chr2R 18693814 99 - 21139217 -ACUCUGCUCGGCACUAG------AGUUAAUCGAAAGAGCCGCCAUAAAUGAAUGAGUUUAUUUA-UUUUUUGGAAUAGUAUUCUUUAUGGCUGCAAUCUGGACGAA -.......(((...((((------(.....((....))((.((((((((.(((((...((((((.-.......))))))))))))))))))).))..))))).))). ( -27.40, z-score = -2.49, R) >droEre2.scaffold_4845 20093242 84 - 22589142 -----------------------ACUCUACUCGAUAGAGCCGCCAUAAAUGAAUGAGUUUAUUUAUUUUUUUGGAAUAGUAUUCUUUAUGGUCGCAAUCAGGUCGAA -----------------------.......(((((.((((.((((((((.(((((..((((..........))))....))))))))))))).))..))..))))). ( -19.00, z-score = -1.67, R) >droAna3.scaffold_13266 15068830 97 - 19884421 -ACUCCUCUUGCCAUUUA---------AACUAAUGUCUGCGAUCAUAAAUGAAUGAUUUUAUUUAUUUUUUUCGCAUAUUAUUCUUUAUGGCUUUAAUCGCGUCGAA -.........(((((...---------...(((((..(((((....(((((((((....)))))))))...))))))))))......)))))............... ( -18.70, z-score = -2.80, R) >dp4.chr3 4211052 96 - 19779522 AACCCUAGCUUCCAGUUA---------ACAUCGUCAGUGGGGCCAUAAAUGAAUGAUUCUAAUU-UAUUUUUUGAAUAUUGUUCUUUAUGGUUCACGUCGCGUGAA- .....((((.....))))---------.((.(((..(((((.(((((((.(((..((....(((-((.....)))))))..)))))))))))))))...)))))..- ( -21.00, z-score = -1.36, R) >droPer1.super_2 4397996 96 - 9036312 AACCCUAGCUUCCAGUUA---------ACAUCGUCAAUGGGGCUAUAAAUGAAUGAUUCUAAUU-UAUUUUUUGAAUAUUAUUCUUUAUGGUUCACGUCGCGUGAA- .....((((.....))))---------.((.(((..(((((((((((((.(((((((....(((-((.....)))))))))))))))))))))).))).)))))..- ( -19.60, z-score = -1.53, R) >consensus _ACUCUGCUCAGCACUAG_________AAAUCGAAAGAGCCGCCAUAAAUGAAUGAGUUUAUUUA_UUUUUUGGAAUAGUAUUCUUUAUGGCCGCAAUCUGGUCGAA ......................................((.((((((((.(((((........................))))))))))))).))............ (-10.98 = -10.57 + -0.40)

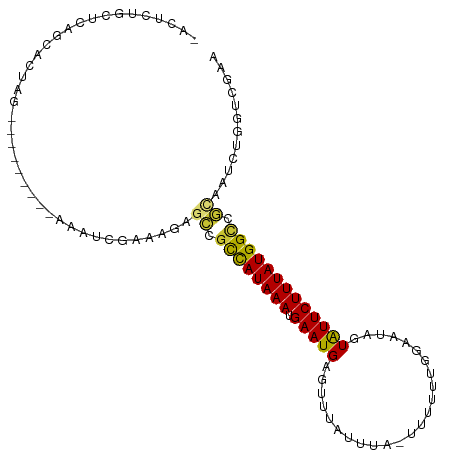
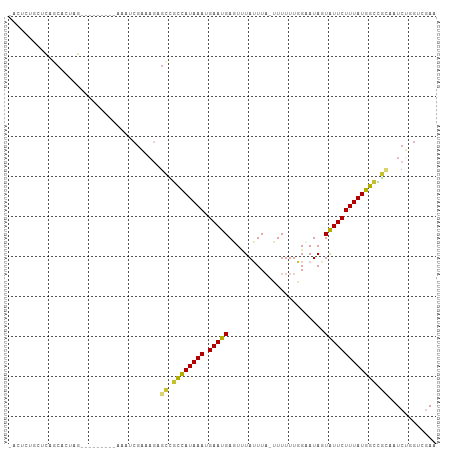
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:19 2011