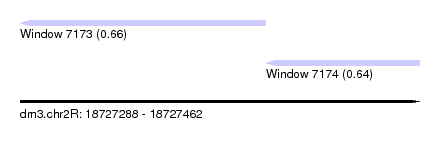
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,727,288 – 18,727,462 |
| Length | 174 |
| Max. P | 0.658607 |

| Location | 18,727,288 – 18,727,395 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 136 |
| Reading direction | reverse |
| Mean pairwise identity | 68.53 |
| Shannon entropy | 0.52893 |
| G+C content | 0.39759 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -13.36 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
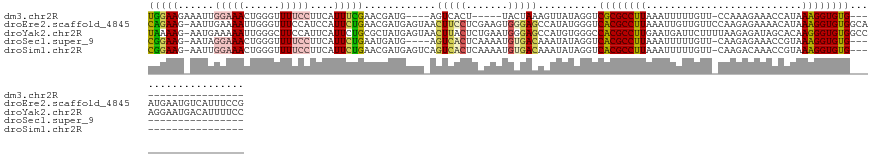
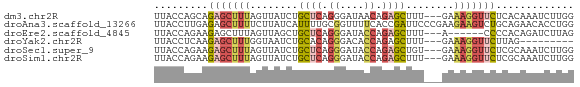
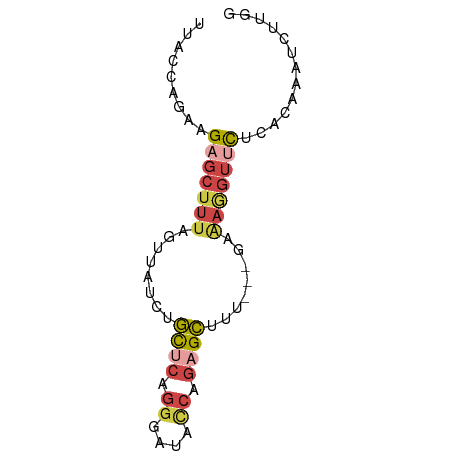
>dm3.chr2R 18727288 107 - 21146708 UGGAAGAAAUUGGAAACUGGGUUUUCCUUCAUUUCGAACGAUG----AGUCACU-----UACUAAAGUUAUAGGUCGCGCCUUAAAUUUUUGUU-CCAAAGAAACCAUAAAGGUGUG------------------- .((((((....(....)....))))))............(((.----.((.(((-----(....)))).))..)))(((((((...(((((...-....))))).....))))))).------------------- ( -19.50, z-score = 0.59, R) >droEre2.scaffold_4845 20092828 135 - 22589142 CAGAAG-AAUUGAAAAUUGGGUUUCCAUCCAUUCUGAACGAUGAGUAACUUCCUCGAAGUGGGAGCCAUAUGGGUCACGCCUUAAAUUGUUGUUCCAAGAGAAAACAUAAAGGUGUGGCAAUGAAUGUCAUUUCCG .....(-((.(((......(((((((....((((........)))).(((((...))))))))))))......((((((((((....((((.(((.....)))))))..))))))))))........))).))).. ( -36.00, z-score = -1.25, R) >droYak2.chr2R 18693383 135 - 21139217 UAAAAG-AAUGAAAAAUUGGGCUUCCAUUCAUUCUGCGCUAUGAGUAACUUACUCUGAAUGGGAGCCAUGUGGGCCACGCCUUGAAUGAUUCUUUUAAGAGAUAGCACAAGGGUGUGGCCAGGAAUGACAUUUUCC ....((-((((........((((((((((((...........(((((...))))))))))))))))).....(((((((((((...((..((((....))))...))..)))))))))))........)))))).. ( -49.40, z-score = -4.31, R) >droSec1.super_9 2027743 111 - 3197100 CGGAAG-AAUAGGAAACUGGGUUUUCCUUCAUUCUGAAUGAUG----AGUCACUCAAAAUGUGACAAAUAUAGGUCACGCCUUAAAUUUUUGUU-CAAGAGAAACCGUAAAGGUGUG------------------- .(((((-(.(((....)))..)))))).(((((......))))----)(((((.......)))))..........((((((((.........((-(....)))......))))))))------------------- ( -28.96, z-score = -2.08, R) >droSim1.chr2R 17323024 115 - 19596830 CGGAAG-AAUUGGAAACUGGGUUUUCCUUCAUUCUGAACGAUGAGUCAGUCACUCAAAAUGUGACAAAUAUAGGUCACGCCUUAAAUUUUUGUU-CAAGACAAACCGUAAAGGUGUG------------------- .(((((-(...(....)....))))))((((((......))))))...(((((.......)))))..........((((((((.....((((((-...)))))).....))))))))------------------- ( -29.40, z-score = -1.55, R) >consensus CGGAAG_AAUUGGAAACUGGGUUUUCCUUCAUUCUGAACGAUGAGU_AGUCACUCAAAAUGGGACAAAUAUAGGUCACGCCUUAAAUUUUUGUU_CAAGAGAAACCAUAAAGGUGUG___________________ (((((......(((((......)))))....)))))............(((((.......)))))..........((((((((...(((((........))))).....))))))))................... (-13.36 = -14.32 + 0.96)
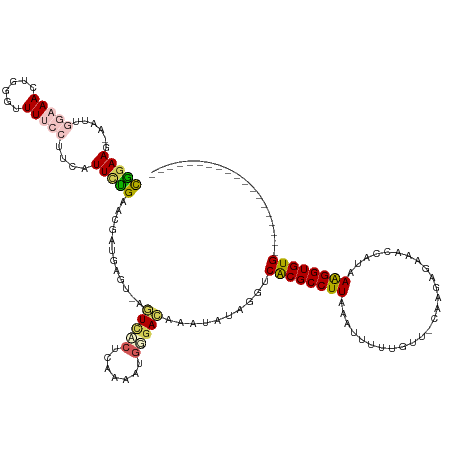
| Location | 18,727,395 – 18,727,462 |
|---|---|
| Length | 67 |
| Sequences | 6 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.56802 |
| G+C content | 0.44144 |
| Mean single sequence MFE | -16.85 |
| Consensus MFE | -7.22 |
| Energy contribution | -8.34 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18727395 67 - 21146708 UUACCAGCAGAGCUUUAGUUAUCUGCUCAGGGAUAACAGAGCUUU---GAAAGGUUCUCACAAAUCUUGG ...((..(((((((((.(((((((......)))))))))))))))---).((((((......)))))))) ( -21.00, z-score = -2.75, R) >droAna3.scaffold_13266 15068550 70 - 19884421 UUACCUUGAGAGCUUUUCUUAUCAUUUUGCGGUUUUCACCGAUUCCCGAAGAAGUCUGCAGAACACCUGG ...((.((((((...))))))...((((((((.((((..((.....))..)))).)))))))).....)) ( -14.00, z-score = -0.80, R) >droEre2.scaffold_4845 20092963 61 - 22589142 UUACCAGAAGAGCUUUAGUUAGCUGCUCAGGGAUACCAGAGCUUU---A------CCCCACAGAUCUUAG .......(((((((......))).((((.((....)).))))...---.------.........)))).. ( -12.40, z-score = -0.72, R) >droYak2.chr2R 18693518 58 - 21139217 UUACCUCAAGAGCUUUGGUAAUCUGCACAGGGACACCAGAGCUUU---GAAAGGUUCUUAG--------- ..((((..(((((((((((..(((....)))...)))))))))))---...))))......--------- ( -20.40, z-score = -3.10, R) >droSec1.super_9 2027854 67 - 3197100 UUACCAGAAGAGCUUUAGUUAUCUGCUCAGGGAUACCAGAGCUGU---GAAAGGUUCUCGCAAAUCUUGG ...(((((((((((((..((((..((((.((....)).)))).))---))))))))))......)).))) ( -17.10, z-score = -0.60, R) >droSim1.chr2R 17323139 67 - 19596830 UUACCAGAAGAGCUUUAGUUAUCUGCUCAGGGAUACCAGAGCUUU---GAAAGGUUCUCGCAAAUCUUGG ...(((((((((((((..(((...((((.((....)).))))..)---))))))))))......)).))) ( -16.20, z-score = -0.48, R) >consensus UUACCAGAAGAGCUUUAGUUAUCUGCUCAGGGAUACCAGAGCUUU___GAAAGGUUCUCACAAAUCUUGG .........(((((((........((((.((....)).))))........)))))))............. ( -7.22 = -8.34 + 1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:18 2011