
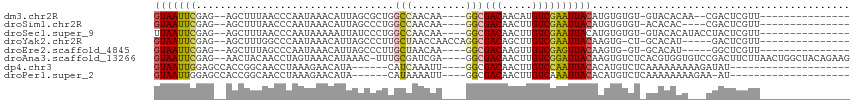
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,697,198 – 18,697,290 |
| Length | 92 |
| Max. P | 0.839806 |

| Location | 18,697,198 – 18,697,290 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.49 |
| Shannon entropy | 0.61199 |
| G+C content | 0.42002 |
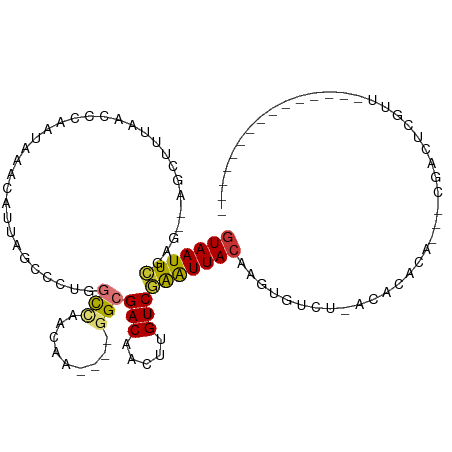
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -9.01 |
| Energy contribution | -9.04 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18697198 92 + 21146708 GUAAUUCGAG--AGCUUUAACCCAAUAAACAUUAGCGCUGGCCAACAA----GGCGACAACAUGUCGAAUUACAUGUGUGU-GUACACAA--CGACUCGUU--------------- (((((((((.--((((.................)))....(((.....----))).......).))))))))).(((((..-..))))).--.........--------------- ( -20.93, z-score = -0.79, R) >droSim1.chr2R 17292559 90 + 19596830 GUAAUUCGAG--AGCUUUAACCCAAUAAACAUUAGCCCUGGCCAACAA----GGCGACAACUUGUCGAAUUACAUGUGUGU-ACACAC----CGACUCGUU--------------- (((((((...--......................(((.((.....)).----)))(((.....)))))))))).((((...-.)))).----.........--------------- ( -20.10, z-score = -1.60, R) >droSec1.super_9 1998543 94 + 3197100 UUAAUUCGAG--AGCUUUAACCCAAUAAAAAUUAUCCCUGGCCAACAA----GGCGACAACUUGUCGAAUUACAUGUGUGU-GUACACAUACCUACUCGUU--------------- .((((((...--............................(((.....----)))(((.....)))))))))..((((((.-...))))))..........--------------- ( -16.70, z-score = -0.82, R) >droYak2.chr2R 16163751 92 + 21139217 GUAAUUCGAG--AGCUUUGGCCCAAUAAACAUUAGCCCUUGCUAACCAACCAGGCGACAGCUUGUCGAAUUACAAGUG-CU-GCACAU-----GACUCGUU--------------- ((((((((((--((((.(.(((.........(((((....))))).......))).).))))).)))))))))..(((-..-.)))..-----........--------------- ( -25.69, z-score = -2.42, R) >droEre2.scaffold_4845 20060588 88 + 22589142 GUAAUUCGAG--AGCUUUAGCCCAAUAAACAUUAGCCCUUGCUAACAA----GGCGACAAGUUGUCGAGUUACAAGUG-GU-GCACAU-----GGCUCGUU--------------- (((((((((.--(((((..(((.........(((((....)))))...----)))...))))).))))))))).(((.-((-....))-----.)))....--------------- ( -25.00, z-score = -1.81, R) >droAna3.scaffold_13266 15041317 109 + 19884421 GUAAUUCGAG--AACUACAACCUAGUAAACAUAAAC-UUUGCGAUCGA----GGCGACAACUUGUCGGAUUACAAGUGUCUCACGUGGUGUCCGACUUCUUAACUGGCUACAGAAG .....(((.(--((((((......(((((.......-)))))....((----(((....((((((......)))))))))))..))))).)))))........(((....)))... ( -23.50, z-score = 0.10, R) >dp4.chr3 4186791 86 + 19779522 GUAAUUGGAGCCACCGGCAACCUAAAGAACAUA------CAUCAAAUU----GGCGACAACUUGUCCAAUUACACAUGUCUCAAAAAAAAAGAUAU-------------------- ((((((((((((...)))...............------...(((.((----(....))).))))))))))))..((((((.........))))))-------------------- ( -19.60, z-score = -3.59, R) >droPer1.super_2 4373703 85 + 9036312 GUAAUUGGAGCCACCGGCAACCUAAAGAACAUA------CAUAAAAUU----GGCGACAACUUGUCAAAUUACACAUGUCUCAAAAAAAAGAA-AU-------------------- ((((((...((((..(....).....(......------).......)----)))(((.....))).))))))....................-..-------------------- ( -11.10, z-score = -0.51, R) >consensus GUAAUUCGAG__AGCUUUAACCCAAUAAACAUUAGCCCUGGCCAACAA____GGCGACAACUUGUCGAAUUACAAGUGUCU_ACACACA___CGACUCGUU_______________ (((((((.................................(((.........)))(((.....))))))))))........................................... ( -9.01 = -9.04 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:15 2011