
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,693,863 – 18,693,932 |
| Length | 69 |
| Max. P | 0.622706 |

| Location | 18,693,863 – 18,693,932 |
|---|---|
| Length | 69 |
| Sequences | 9 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 64.19 |
| Shannon entropy | 0.72398 |
| G+C content | 0.47423 |
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -6.55 |
| Energy contribution | -6.91 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
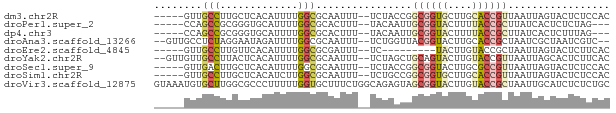
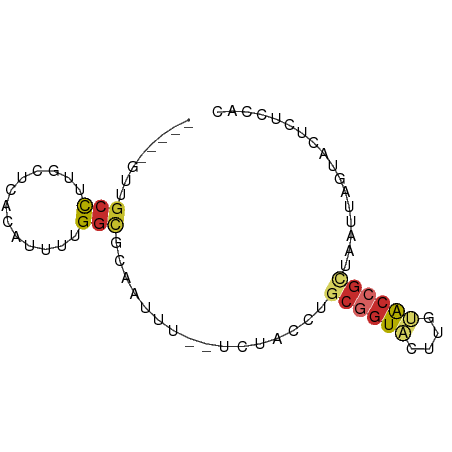
>dm3.chr2R 18693863 69 - 21146708 -----GUUGCCUUGCUCACAUUUUGGCGCAAUUU--UCUACCGGCGGUGCUUGCACCGUUAAUUAGUACUCUCCAC -----(((((...(((........))))))))..--......(((((((....)))))))................ ( -14.40, z-score = -0.54, R) >droPer1.super_2 4368483 66 - 9036312 -----CCAGCCGCGGGUGCAUUUUGGCGCACUUU--UACAAUUGCGGUACUUUUACCGCUUAUCACUCUCUAG--- -----........((((((........)))))).--.......((((((....))))))..............--- ( -14.50, z-score = -0.76, R) >dp4.chr3 15604794 66 + 19779522 -----CCAGCCGCGGGUGCAUUUUGGCGCACUUU--UACAAUUGCGGUACUUUUACCGCUUAUCACUCUUUAG--- -----........((((((........)))))).--.......((((((....))))))..............--- ( -14.50, z-score = -0.80, R) >droAna3.scaffold_13266 15038096 70 - 19884421 --GUUGCCUCUAGGAAUAGAUUUUGGCGCAAUUU--UCUGGUUACGGUACUUGCACCGCUAAUCGCUAAUCGUC-- --((.(((((((....))))....))))).....--...(((((((((......)))).)))))..........-- ( -13.70, z-score = 0.15, R) >droEre2.scaffold_4845 20057268 60 - 22589142 -----GUUGCCUUGUUCACAUUUUGGCGCGAUUU--UC---------UACUUGUACCGCUAAUUAGUACUCUUCAC -----...(((.((....))....)))(((....--..---------.........)))................. ( -6.06, z-score = 0.35, R) >droYak2.chr2R 16160424 72 - 21139217 --GUUGUUGCCUUACUCACAUUUUGGCGCAAUUU--UCUAGCUGCAGUACUUGUACCGUUAAUUAGCACUCUUCAC --(((((.(((.............))))))))..--....(((((.(((....))).)).....)))......... ( -9.92, z-score = 0.42, R) >droSec1.super_9 1995220 69 - 3197100 -----GUUGACUUGCUCACAUUUUGGCGCAAUUU--UCUACCGGCGGUACUUGCGCCGUUAAUUAGUACUCUCCAC -----(..((..((((.......((((((((...--..((((...)))).))))))))......)))).))..).. ( -14.52, z-score = -0.87, R) >droSim1.chr2R 17289079 69 - 19596830 -----GUUGCCUUGCUCACAUCUUGGCGCAAUUU--UCUGCCGGCGGUGCUUGCACCGUUAAUUAGUACUCUCCAC -----.......((((........((((......--..))))(((((((....)))))))....))))........ ( -15.70, z-score = -0.46, R) >droVir3.scaffold_12875 13942244 76 - 20611582 GUAAAUGUGCUUGGCGCCCUUUUUGGUGCUUUCUGGCAGAGUAGCGGUACUUGUACCGCUAAUUGCAUCUCUCUGC ((((.(.((((.((((((......))))))....)))).).((((((((....)))))))).)))).......... ( -27.90, z-score = -3.20, R) >consensus _____GUUGCCUUGCUCACAUUUUGGCGCAAUUU__UCUACCUGCGGUACUUGUACCGCUAAUUAGUACUCUCCAC ........(((.............)))................((((((....))))))................. ( -6.55 = -6.91 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:14 2011