
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,691,338 – 18,691,411 |
| Length | 73 |
| Max. P | 0.985667 |

| Location | 18,691,338 – 18,691,411 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 73.86 |
| Shannon entropy | 0.55155 |
| G+C content | 0.45916 |
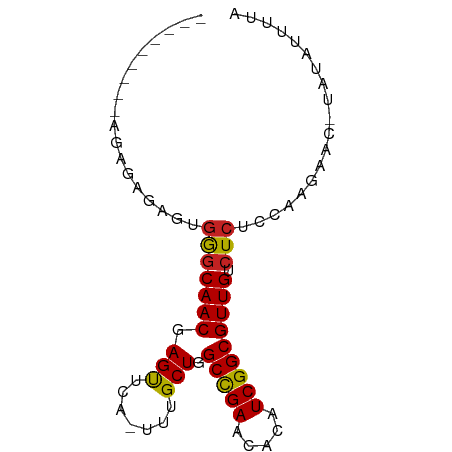
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.65 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
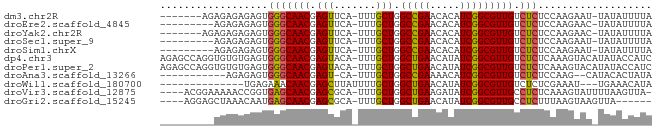
>dm3.chr2R 18691338 73 - 21146708 -------AGAGAGAGAGUGGGCAACGAGUUCA-UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAU-UAUAUUUUA -------.((((((((((((((.....)))))-)))....(((((.....)))))....)))))).......-......... ( -22.50, z-score = -2.19, R) >droEre2.scaffold_4845 20054798 71 - 22589142 ---------AGAGAGAGUGGGCAACGAGUUCA-UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAC-UAUAUUUUA ---------...(.(((.(((((((.(((...-...))).(((((.....))))))))))))))))......-......... ( -20.80, z-score = -1.68, R) >droYak2.chr2R 16157879 73 - 21139217 -------AGAGAGAGAGUGGGCAACGAGUUCA-UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAC-UAUAUUUUA -------.((((((((((((((.....)))))-)))....(((((.....)))))....)))))).......-......... ( -22.50, z-score = -1.96, R) >droSec1.super_9 1992727 71 - 3197100 ---------AGAGAGAGUGGGCAACGAGUUCA-UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAU-UAUAUUUUA ---------...(.(((.(((((((.(((...-...))).(((((.....))))))))))))))))......-......... ( -20.80, z-score = -1.88, R) >droSim1.chrX 16221654 71 - 17042790 ---------AGAGAGAGUGGGCAACGAGUUCA-UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAU-UAUAUUUUA ---------...(.(((.(((((((.(((...-...))).(((((.....))))))))))))))))......-......... ( -20.80, z-score = -1.88, R) >dp4.chr3 15602396 81 + 19779522 AGAGCCAGGUGUGUGAGUGGGCAACGAGUACA-UUUGCUGGCUGAACAUAUCGGCGUUGUCUCUCAAAGUACAUAUACCAUC .......((((((((((.(((((((.((((..-..)))).(((((.....)))))))))))))))).......))))))... ( -24.71, z-score = -1.28, R) >droPer1.super_2 4366086 81 - 9036312 AGAGCCAGGUGUGUGAGUGGGCAACGAGUACA-UUUGCUGGCUGAACAUAUCGGCGUUGUCUCUCAAAGUACAUAUACCAUC .......((((((((((.(((((((.((((..-..)))).(((((.....)))))))))))))))).......))))))... ( -24.71, z-score = -1.28, R) >droAna3.scaffold_13266 15035750 67 - 19884421 -----------AGAGAGUGGGCAACGAGU-CA-UUUGCUGGCCGAAAACAUCGGCGUUGUCUCUCCAAG--CAUACACUAUA -----------.(.(((.(((((((.(((-..-...))).(((((.....))))))))))))))))...--........... ( -19.80, z-score = -1.49, R) >droWil1.scaffold_180700 2787395 65 + 6630534 --------------UGAGAAACAACGAGCUUAUUUUGCUGGCUGAACAUAUCGGCGUUGUCUCUCGAAAU---UGAAACAUA --------------.((((.(((((.(((.......))).(((((.....)))))))))).)))).....---......... ( -19.60, z-score = -3.54, R) >droVir3.scaffold_12875 13939824 76 - 20611582 ----ACGGAAAAACCGGUGAGCAACGAGCGCA-UUUGCUGGCUGAAGAUAUCGGCGUUGCCUCUCAAAGUAUUUUAAGUUA- ----.(((.....)))(.(((((((.((((..-..)))).(((((.....))))))))).))).)................- ( -19.50, z-score = -0.35, R) >droGri2.scaffold_15245 1029082 71 + 18325388 ----AGGAGCUAAACAAUGAGCAACGAGCGCA-UUUGCUGGCUGAACAUAUCGGCGUUGCCUCUUUAAGUAAGUUA------ ----...((((.......(((((((.((((..-..)))).(((((.....))))))))).)))........)))).------ ( -16.86, z-score = 0.21, R) >consensus _________AGAGAGAGUGGGCAACGAGUUCA_UUUGCUGGCCGAACACAUCGGCGUUGUCUCUCCAAGAAC_UAUAUUUUA ..................(((((((.(((.......))).(((((.....))))))))).)))................... (-14.12 = -13.65 + -0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:13 2011