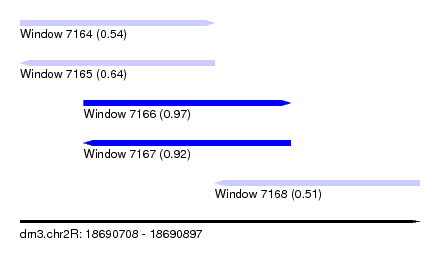
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,690,708 – 18,690,897 |
| Length | 189 |
| Max. P | 0.972742 |

| Location | 18,690,708 – 18,690,800 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.40182 |
| G+C content | 0.50102 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.06 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
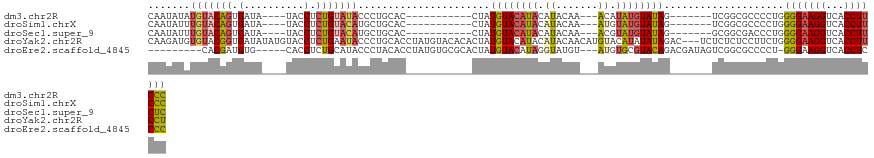
>dm3.chr2R 18690708 92 + 21146708 AAGCGGAAGAGAAGUAACUGACCUGCGAUCGGGAAGGUGACCUUCCCCAGG------GGCGCCGACUAUACAUAUGU---UUGUAUGUAUGUACAUAG-GUG---------- ...(....)............((((.....(((((((...)))))))))))------..((((.((.((((((((..---..)))))))))).....)-)))---------- ( -26.80, z-score = -0.93, R) >droSim1.chrX 16221022 92 + 17042790 AAGCGGAAGAGAAGUAACUGACCUGCGAUCGGGAAGGUGACCUUCCCCAGG------GGCGCCGACUAUACAUACAU---UUGUAUGUAUGUACAUAG-GUG---------- ...(....)............((((.....(((((((...)))))))))))------..((((.....(((((((((---....)))))))))....)-)))---------- ( -29.30, z-score = -1.86, R) >droSec1.super_9 1992093 92 + 3197100 AAGCGGAAGAGAAGUAACUGACCUGCGAUCGAGAAGGUGACCUUCCCCAGG------GUCGCCGCCUAUACAUACGU---UUGUAUGUAUGUACAUAG-GUG---------- ..((((.((........))...)))).........((((((((......))------))))))((((((((((((((---....)))))))))..)))-)).---------- ( -32.50, z-score = -3.25, R) >droYak2.chr2R 16157351 110 + 21139217 AAGCGGAAGAGGAGUAACUGACCUGCGAUCAGGAAGGUGACCUUCCCCAGAAGG--AGAGAGAGUCUAUAUAUGUACAUGUUGUAUGUAUGUACAUAGUGUGUACAUAGGUG ...(....).(((.(..((..(((.(.....((((((...))))))...).)))--..))..).)))...((((((((..((((((....)))))..)..)))))))).... ( -27.50, z-score = -0.82, R) >droEre2.scaffold_4845 20054172 109 + 22589142 AAGCGGAAGAGGAGCAACUGACCUGCGAUCGGGGAGGUGACCUUCCCAGGGGCGCCGACUAUCGUCUGUACGCACAU---ACAUACCUAUGUACAUAGUGCGCACAUAGGUG ..(((..(((.(((((.......)))..(((((((((...))))))).((....))))...)).)))...)))....---...(((((((((.(.......).))))))))) ( -34.60, z-score = -0.22, R) >consensus AAGCGGAAGAGAAGUAACUGACCUGCGAUCGGGAAGGUGACCUUCCCCAGG______GGCGCCGACUAUACAUACAU___UUGUAUGUAUGUACAUAG_GUG__________ ..((((...((......))...))))....(((((((...)))))))...................((((((((((.....))))))))))..................... (-18.10 = -18.06 + -0.04)

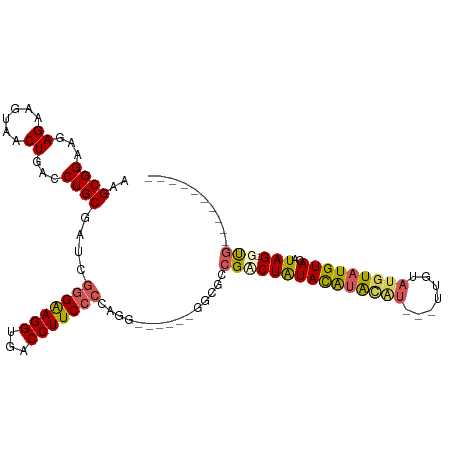
| Location | 18,690,708 – 18,690,800 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.40182 |
| G+C content | 0.50102 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18690708 92 - 21146708 ----------CAC-CUAUGUACAUACAUACAA---ACAUAUGUAUAGUCGGCGCC------CCUGGGGAAGGUCACCUUCCCGAUCGCAGGUCAGUUACUUCUCUUCCGCUU ----------.((-((.((.(((((((((...---...))))))).))))(((..------..(.(((((((...))))))).).))))))).................... ( -22.40, z-score = -0.93, R) >droSim1.chrX 16221022 92 - 17042790 ----------CAC-CUAUGUACAUACAUACAA---AUGUAUGUAUAGUCGGCGCC------CCUGGGGAAGGUCACCUUCCCGAUCGCAGGUCAGUUACUUCUCUUCCGCUU ----------.((-((.(((((((((((....---)))))))))))....(((..------..(.(((((((...))))))).).))))))).................... ( -28.80, z-score = -2.71, R) >droSec1.super_9 1992093 92 - 3197100 ----------CAC-CUAUGUACAUACAUACAA---ACGUAUGUAUAGGCGGCGAC------CCUGGGGAAGGUCACCUUCUCGAUCGCAGGUCAGUUACUUCUCUUCCGCUU ----------...-...(((((((((......---..)))))))))(((((.(((------(.(((((((((...))))))).....))))))((......))...))))). ( -27.10, z-score = -1.95, R) >droYak2.chr2R 16157351 110 - 21139217 CACCUAUGUACACACUAUGUACAUACAUACAACAUGUACAUAUAUAGACUCUCUCU--CCUUCUGGGGAAGGUCACCUUCCUGAUCGCAGGUCAGUUACUCCUCUUCCGCUU ....((((((((...(((((....))))).....))))))))....(((.((.(((--((....))))))))))......(((((.....)))))................. ( -25.30, z-score = -1.96, R) >droEre2.scaffold_4845 20054172 109 - 22589142 CACCUAUGUGCGCACUAUGUACAUAGGUAUGU---AUGUGCGUACAGACGAUAGUCGGCGCCCCUGGGAAGGUCACCUCCCCGAUCGCAGGUCAGUUGCUCCUCUUCCGCUU .((((.((((((((((((((((....))))))---).)))))))))(((....))).(((....((((.(((...))).))))..))))))).................... ( -36.40, z-score = -1.02, R) >consensus __________CAC_CUAUGUACAUACAUACAA___AUGUAUGUAUAGACGGCGCC______CCUGGGGAAGGUCACCUUCCCGAUCGCAGGUCAGUUACUUCUCUUCCGCUU .................((((((((.............))))))))...................(((((((...)))))))....((.((..((......))...)))).. (-15.36 = -15.52 + 0.16)

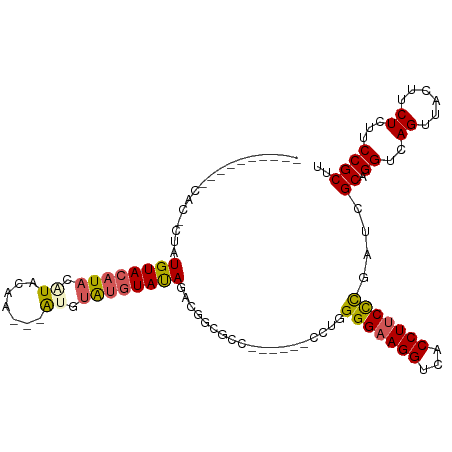
| Location | 18,690,738 – 18,690,836 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 69.69 |
| Shannon entropy | 0.49256 |
| G+C content | 0.46320 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.16 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18690738 98 + 21146708 GGGAAGGUGACCUUCCCCAGGGGCGCCGA-------CUAUACAUAUGU---UUGUAUGUAUGUACAUAG-----------GUGCAGGGUAUACAGAGGUA----UAUCACUGUACAUAUAUUG (((((((...)))))))(((..(((((.(-------(.((((((((..---..)))))))))).....)-----------))))..(((((((....)))----)))).)))........... ( -34.90, z-score = -2.13, R) >droSim1.chrX 16221052 98 + 17042790 GGGAAGGUGACCUUCCCCAGGGGCGCCGA-------CUAUACAUACAU---UUGUAUGUAUGUACAUAG-----------GUGCAGCAUGUACAGAGGUA----UAUCACUGUACAAAUAUUG (((((((...)))))))..(..(((((..-------...(((((((((---....)))))))))....)-----------))))..).(((((((.((..----..)).)))))))....... ( -37.40, z-score = -3.55, R) >droSec1.super_9 1992123 98 + 3197100 GAGAAGGUGACCUUCCCCAGGGUCGCCGC-------CUAUACAUACGU---UUGUAUGUAUGUACAUAG-----------GUGCAGCAUGUACAGAGGUA----UAUCACUGUACAAAUAUUG .....((((((((......))))))))((-------((((((((((((---....)))))))))..)))-----------))......(((((((.((..----..)).)))))))....... ( -36.80, z-score = -3.78, R) >droYak2.chr2R 16157381 120 + 21139217 AGGAAGGUGACCUUCCCCAGAAGGAGAGAGA---GUCUAUAUAUGUACAUGUUGUAUGUAUGUACAUAGUGUGUACAUAGGUGCAGGGUAUUCAGAGGUACAUAUAUCACCGUACACAUCUUG .(...((((((((((....))))).......---.....(((((((((((.((((((.(((((((((...))))))))).)))))).))........))))))))))))))...)........ ( -38.60, z-score = -2.29, R) >droEre2.scaffold_4845 20054202 105 + 22589142 GGGGAGGUGACCUUCCC-AGGGGCGCCGACUAUCGUCUGUACGCACAU---ACAUACCUAUGUACAUAGUGCGCACAUAGGUGUAGGGUAUGCAGAGGUG-----CACAUCGUG--------- (((((((...)))))))-.((.((((((.....).((((((..(...(---((..((((((((.(.......).)))))))))))..)..))))))))))-----)...))...--------- ( -39.50, z-score = -1.15, R) >consensus GGGAAGGUGACCUUCCCCAGGGGCGCCGA_______CUAUACAUACAU___UUGUAUGUAUGUACAUAG___________GUGCAGGGUAUACAGAGGUA____UAUCACUGUACAAAUAUUG (((((((...))))))).....................(((((((((.....)))))))))((((...............((((.....))))...(((.........)))))))........ (-16.24 = -16.16 + -0.08)
| Location | 18,690,738 – 18,690,836 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 69.69 |
| Shannon entropy | 0.49256 |
| G+C content | 0.46320 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -13.88 |
| Energy contribution | -14.84 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
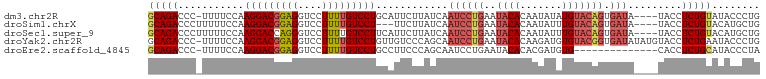
>dm3.chr2R 18690738 98 - 21146708 CAAUAUAUGUACAGUGAUA----UACCUCUGUAUACCCUGCAC-----------CUAUGUACAUACAUACAA---ACAUAUGUAUAG-------UCGGCGCCCCUGGGGAAGGUCACCUUCCC .......(((((((.(...----...).)))))))....((.(-----------(..((((((((.......---...)))))))).-------..)).)).....(((((((...))))))) ( -26.40, z-score = -1.13, R) >droSim1.chrX 16221052 98 - 17042790 CAAUAUUUGUACAGUGAUA----UACCUCUGUACAUGCUGCAC-----------CUAUGUACAUACAUACAA---AUGUAUGUAUAG-------UCGGCGCCCCUGGGGAAGGUCACCUUCCC .......(((((((.(...----...).)))))))....((.(-----------(..(((((((((((....---))))))))))).-------..)).)).....(((((((...))))))) ( -35.00, z-score = -3.53, R) >droSec1.super_9 1992123 98 - 3197100 CAAUAUUUGUACAGUGAUA----UACCUCUGUACAUGCUGCAC-----------CUAUGUACAUACAUACAA---ACGUAUGUAUAG-------GCGGCGACCCUGGGGAAGGUCACCUUCUC .......(((((((.(...----...).)))))))((((((..-----------...(((((((((......---..))))))))).-------))))))......(((((((...))))))) ( -33.60, z-score = -3.23, R) >droYak2.chr2R 16157381 120 - 21139217 CAAGAUGUGUACGGUGAUAUAUGUACCUCUGAAUACCCUGCACCUAUGUACACACUAUGUACAUACAUACAACAUGUACAUAUAUAGAC---UCUCUCUCCUUCUGGGGAAGGUCACCUUCCU ....((((((((((((......(((........)))....))))((((((((.....))))))))..........))))))))...(((---.((.(((((....))))))))))........ ( -31.50, z-score = -1.71, R) >droEre2.scaffold_4845 20054202 105 - 22589142 ---------CACGAUGUG-----CACCUCUGCAUACCCUACACCUAUGUGCGCACUAUGUACAUAGGUAUGU---AUGUGCGUACAGACGAUAGUCGGCGCCCCU-GGGAAGGUCACCUCCCC ---------...((.(((-----.((((((((((((.....(((((((((((.....)))))))))))..))---))))((((...(((....))).))))....-.)).))))))).))... ( -37.60, z-score = -1.86, R) >consensus CAAUAUUUGUACAGUGAUA____UACCUCUGUAUACCCUGCAC___________CUAUGUACAUACAUACAA___AUGUAUGUAUAG_______UCGGCGCCCCUGGGGAAGGUCACCUUCCC .......(((((((.(..........).)))))))......................((((((((.............))))))))....................(((((((...))))))) (-13.88 = -14.84 + 0.96)
| Location | 18,690,800 – 18,690,897 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.58 |
| Shannon entropy | 0.30523 |
| G+C content | 0.44907 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -13.66 |
| Energy contribution | -15.10 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
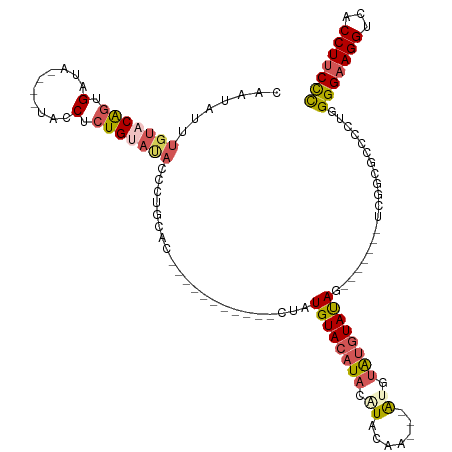
>dm3.chr2R 18690800 97 - 21146708 GCAGACCC-UUUUCCAAGGACGGAGGUCCUUUUGUCCUGCAUUCUUAUCAAUCCUGAAUACACAAUAUAUGUACAGUGAUA----UACCUCUGUAUACCCUG .(((..((-((....))))((((((((.....(((((((........(((....))).((((.......))))))).))))----.)))))))).....))) ( -19.90, z-score = -1.31, R) >droSim1.chrX 16221114 95 - 17042790 GCAGACCCUUUUUCCAAGGACGGAGGUCCUUUUGUCCU---UUCUUAUCAAUCCUGAAUACACAAUAUUUGUACAGUGAUA----UACCUCUGUACAUGCUG (((............((((((((((....)))))))))---)...........................(((((((.(...----...).)))))))))).. ( -21.50, z-score = -2.07, R) >droSec1.super_9 1992185 98 - 3197100 GCAGACCCUUUUUCCAAGGACCAGGGUCCUUUUGUCCUUCAUUCUUAUCAAUCCUGAAUACACAAUAUUUGUACAGUGAUA----UACCUCUGUACAUGCUG (((((((((...((....))..))))))...((((..((((.............))))...))))....(((((((.(...----...).)))))))))).. ( -22.02, z-score = -2.33, R) >droYak2.chr2R 16157461 101 - 21139217 GCAGACCC-UUUUCCAAGGACGGAGGUCCUUUUGUCCUGUUGUCCCAGCAAUCCUGAAUACACAAGAUGUGUACGGUGAUAUAUGUACCUCUGAAUACCCUG .(((..((-((....)))).(((((((.(...(((((((......(((.....)))..(((((.....)))))))).))))...).)))))))......))) ( -25.40, z-score = -1.23, R) >droEre2.scaffold_4845 20054281 87 - 22589142 GCAGACCC-UUUUCCAAGGACGGAGGUCCUUUUGUCCUGCCUUCCCAGCAAUCCUGAAUACACACGAUGUG--------------CACCUCUGCAUACCCUA (((((...-.......(((((((((....))))))))).........((((((.((......)).))).))--------------)...)))))........ ( -21.70, z-score = -2.00, R) >consensus GCAGACCC_UUUUCCAAGGACGGAGGUCCUUUUGUCCUGCAUUCUUAUCAAUCCUGAAUACACAAUAUGUGUACAGUGAUA____UACCUCUGUAUACCCUG (((((...........(((((((((....)))))))))............((((((..((((.......))))))).))).........)))))........ (-13.66 = -15.10 + 1.44)
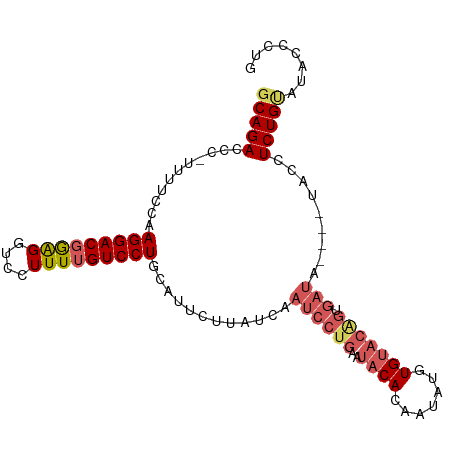
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:13 2011