| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,687,917 – 18,688,025 |
| Length | 108 |
| Max. P | 0.997254 |

| Location | 18,687,917 – 18,688,025 |
|---|---|
| Length | 108 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.38 |
| Shannon entropy | 0.76854 |
| G+C content | 0.48904 |
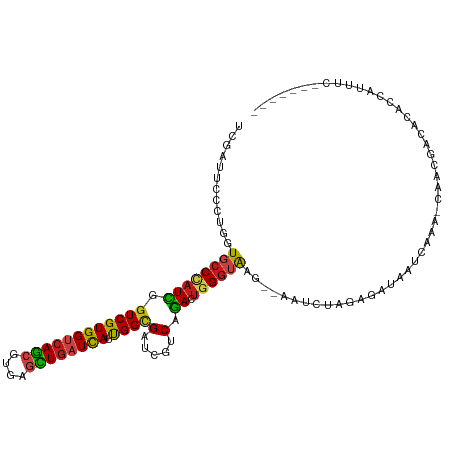
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -18.03 |
| Energy contribution | -20.30 |
| Covariance contribution | 2.27 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
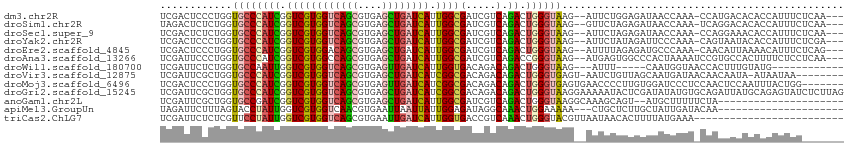
>dm3.chr2R 18687917 108 + 21146708 UCGACUCCCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG--AUUCUGGAGAUAACCAAA-CCAUGACACACCAUUUCUCAA--- .........(((((.......((((((((((((....)))))...((((..(((..((((((.....))--..))))..)))..)))).-))))))).)))))........--- ( -37.00, z-score = -1.59, R) >droSim1.chr2R 17283746 108 + 19596830 UAGACUCUCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG--GUUCUAGAGAUAACCAAA-UCAGGACACACCAUUUCUCAA--- .....(((((((.(((..(((((((((((((((....)))))))))((((....))))))))))....)--)).)))))))...((...-...))................--- ( -41.20, z-score = -3.14, R) >droSec1.super_9 1989258 108 + 3197100 UCGACUCUCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG--AUUCUAGAGAUAACCAAA-CCAGGAAACACCAUUUCUCAA--- .....(((((((((((((...((((((((((((....)))))))))((((....)))))))))))))..--...)))))))........-...(....)............--- ( -38.80, z-score = -2.63, R) >droYak2.chr2R 16154506 108 + 21139217 UCGACUCCCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG--AUUCUAUAGAUUCCCAAA-CAGUAAUACACCAUUUCUCGA--- ((((.....(((((.....((((((((((((((....)))))))....)))))))....((((..(..(--(.((....)).))..)..-))))....)))))....))))--- ( -36.00, z-score = -2.05, R) >droEre2.scaffold_4845 20051338 108 + 22589142 UCGACUCCCUGGUGCCCAUCGGUCGUGGACAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG--AUUUUAGAGAUGCCCAAA-CAACAUUAAAACAUUUCUCAG--- ........(((.((((((.(((((((.((((((....)))).))....)))))))......))))))..--.....(((((((......-...........))))))))))--- ( -30.43, z-score = -0.79, R) >droAna3.scaffold_13266 15032753 109 + 19884421 UCGAUUCCCUGGUGCCCAUCGGUCGUGGCCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACCGGGUAAG--AUGAGUGGCCCACUAAAAUCCGUGCCACUUUUCUCCUCAA--- ..........(((((((...(((((((..((((....))))..)))((((....)))))))))))))((--(.(((((((..((........))))))))).)))))....--- ( -40.20, z-score = -2.12, R) >droWil1.scaffold_180700 2781231 94 - 6630534 UCGAUUCUCUGGUGCCAAUUGGUCGUGGUCAGCGUGAGUUGAUCAUUGGUGACAGACAGACUGGGUAAG---AUUU-----CAAUGGUAACCACUUUGUAUG------------ .........((((((((.(..((((((((((((....)))))))))((....))....)))..)....(---....-----)..)))).)))).........------------ ( -29.10, z-score = -1.45, R) >droVir3.scaffold_12875 13936077 104 + 20611582 UCGAUUCGCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUCGGCGACAGACAGACUGGGUGAGU-AAUCUGUUAGCAAUGAUAACAACAAUA-AUAAUAA-------- .......(((((.(((((((.((((((((((((....))))))))).(....).))).)).)))))((..-..))..)))))................-.......-------- ( -34.30, z-score = -2.18, R) >droMoj3.scaffold_6496 9367576 107 + 26866924 UCGACUCCCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGUUGAUCAUCGGCGACAGACAGACUGGGUGAGUGAACCCCUUGUGGAUCCCUCCAACUCCAAUUUACUGG------- .......((....(((((((.((((((((((((....))))))))).(....).))).)).))))).((((((.......((((....)))).......))))))))------- ( -34.24, z-score = 0.16, R) >droGri2.scaffold_15245 1023557 114 - 18325388 UCGAUUCGCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUCGGCGACAGACAGACUGGGUAAGGAAAAAUACUCGAUAUAUGUGCAGAUUAUGCAGAGUAUCUCUUAG ....(((.((..((((((((.((((((((((((....))))))))).(....).))).)).)))))))))))........(((((.(.((((.....)))).))))))...... ( -39.30, z-score = -1.99, R) >anoGam1.chr2L 9787620 91 + 48795086 UCGAUUCGCUGGUGCCGAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAGGCAAAGCAGU--AUGCUUUUUCUA--------------------- .......(((..((((..(((((((((((((((....)))))))))((((....))))))))))....)))).)))...--............--------------------- ( -38.70, z-score = -3.51, R) >apiMel3.GroupUn 236242255 90 + 399230636 UAGAUUCUUUAGUACCUAUUGGUCGUGGUCAACGUGAAUUAAUUAUUGGAGAUAGGCAAACUGGAAAAA---CUGCUCUUGCUAUUGAUACAA--------------------- ..((((.....(.(((....))))..))))....((.((((((....((((.(((..............---)))))))....)))))).)).--------------------- ( -11.94, z-score = 1.44, R) >triCas2.ChLG7 3374430 89 - 17478683 UCGAUUCUCUCGUUCCUAUUGGUCGUGGUCAGCGUGAAUUGAUCAUUGGUGACCGUCAAACUGGGUACGUUAAUAACACUUUUAUGAAA------------------------- ..........(((.((((((((...((((((.(((((.....))).)).))))))))))..)))).)))....................------------------------- ( -17.70, z-score = 0.47, R) >consensus UCGAUUCCCUGGUGCCCAUCGGUCGUGGUCAGCGUGAGCUGAUCAUUGGCGAUCGUCAGACUGGGUAAG__AAUCUAGAGAUAAUCAAA_CAACGACACACCAUUUC_______ ............((((((((.((((((((((((....)))))))).))))(.....).)).))))))............................................... (-18.03 = -20.30 + 2.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:09 2011