
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,652,995 – 18,653,088 |
| Length | 93 |
| Max. P | 0.549387 |

| Location | 18,652,995 – 18,653,088 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.55286 |
| G+C content | 0.47728 |
| Mean single sequence MFE | -23.04 |
| Consensus MFE | -12.83 |
| Energy contribution | -12.93 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
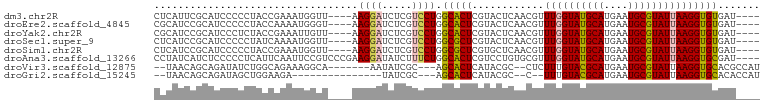
>dm3.chr2R 18652995 93 + 21146708 CUCAUUCGCAUCCCCCUACCGAAAUGGUU----AAGGAUCUCGUCCUGGCACUCGUACUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU---- .....(((((((.......(((....(((----(.((((...))))))))..)))..........((((((((((....))))))))))))))))).---- ( -22.90, z-score = -0.57, R) >droEre2.scaffold_4845 20014536 93 + 22589142 CGCAUCCGCAUCCCCCUACCAAAAUGGGU----AAGGAUCUCGUCCUGGCACUCGUACUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU---- ......((((((...........((((((----.(((((...)))))...)))))).........((((((((((....))))))))))))))))..---- ( -24.40, z-score = -0.40, R) >droYak2.chr2R 16119208 93 + 21139217 CGCAUCCGCAUCCCUCUACCGAAAUUGUU----AAGGAUCUCGUCCUGGCACUCGUACUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU---- ......((((((.......(((...((((----(.((((...))))))))).)))..........((((((((((....))))))))))))))))..---- ( -22.20, z-score = -0.39, R) >droSec1.super_9 1954100 93 + 3197100 CUCAUCCGCAUCCCCCUAUCAAAAUGGUU----AAGGAUCUCGUCCUGGCGCUCGUACUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU---- ......((((((...........(((((.----.(((((...)))))...)).))).........((((((((((....))))))))))))))))..---- ( -21.00, z-score = -0.12, R) >droSim1.chr2R 17249178 93 + 19596830 CUCAUCCGCAUCCCCCUACCGAAAUGGUU----AAGGAUCUCGUCCUGGCGCUCGUGCUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU---- ......((((((..((((((.....))).----.)))....(((...((((....))))..))).((((((((((....))))))))))))))))..---- ( -23.30, z-score = -0.05, R) >droAna3.scaffold_13266 14992059 97 + 19884421 CCUAUCAUCUCCCCCUCAUUCAAUUCCGUCCCGAAGGAUAUCUUUCUGGCACUCGUCCUGUGCGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGCGAU---- ..........................((..(((((((....)))))..((((.......))))..((((((((((....))))))))))))..))..---- ( -24.00, z-score = -1.34, R) >droVir3.scaffold_12875 13888666 87 + 20611582 --UAACAGCAGAUAUCUGGCAGAAAGGCA-------AAUAUCGC---AGCACUCAUACGC--CUCUUUGUACGCAUGAAUGCGUAUUAAGGUGCACGCCAU --.....((.(((((.((.(.....).))-------.)))))))---...........((--.((((.(((((((....))))))).)))).))....... ( -22.80, z-score = -0.88, R) >droGri2.scaffold_15245 976351 77 - 18325388 --UAACAGCAGAUAGCUGGAAGA---------------UAUCGC---AGCACUCAUACGC--C--UUUGUACGCAUGAAUGCGUAUUAAGGUGCACACCAU --.....((.((..((((((...---------------..)).)---)))..))....((--(--((.(((((((....))))))).)))))))....... ( -23.70, z-score = -2.62, R) >consensus CUCAUCCGCAUCCCCCUACCAAAAUGGUU____AAGGAUCUCGUCCUGGCACUCGUACUCAACGUUUGGUAUGCAUGAAUGCGUAUUAAGGUGUGAU____ ..................................((((.....)))).(((((............((((((((((....)))))))))))))))....... (-12.83 = -12.93 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:04 2011