| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,616,387 – 18,616,448 |
| Length | 61 |
| Max. P | 0.628623 |

| Location | 18,616,387 – 18,616,448 |
|---|---|
| Length | 61 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.50802 |
| G+C content | 0.39170 |
| Mean single sequence MFE | -11.45 |
| Consensus MFE | -5.30 |
| Energy contribution | -5.57 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
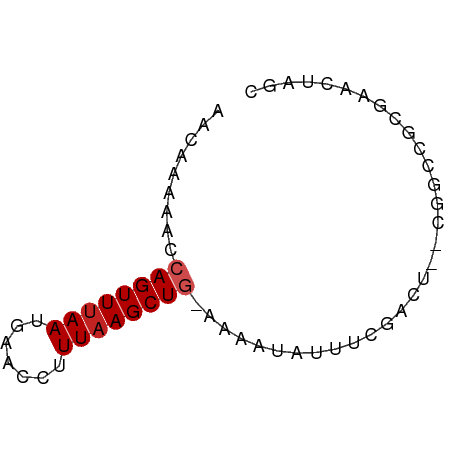
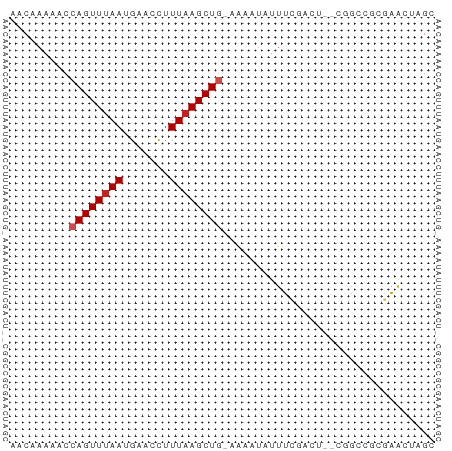
>dm3.chr2R 18616387 61 - 21146708 AACAAAAACCAGUUUAAUGAACCUUUAAGCUG-GAAAUAUUUCGACU--CCACGGCGAACUCGC ........(((((((((.......))))))))-).............--.....(((....))) ( -12.80, z-score = -2.41, R) >droSim1.chr2R 17211812 61 - 19596830 AACAAAAACCAGUUUAAUGAACCUUUAAGCUG-GAAAUAUUUCGACU--CCACAGCGAACUCGC ........(((((((((.......))))))))-).............--.....(((....))) ( -13.00, z-score = -3.15, R) >droSec1.super_9 1918411 61 - 3197100 AACAAAAACCAGUUUAAUGAACCUUUAAGCUG-GAAAUAUUUCGACU--CCACAGCGAACUCGC ........(((((((((.......))))))))-).............--.....(((....))) ( -13.00, z-score = -3.15, R) >droYak2.chr2R 16082151 61 - 21139217 AACAAAAACCAGUUUAAUGAACCUUUAAGCUG-GAAAUAUUUCGACU--CAACGGCGAACUGGC ........(((((((((.......))))))))-)......((((.((--....))))))..... ( -12.60, z-score = -2.16, R) >droEre2.scaffold_4845 19978079 61 - 22589142 AACAAAAACCAGUUUAAUGAACCUUUAAGCUG-AAAAUAUUUCGAAU--CAACAGCGAACUCGC .........((((((((.......))))))))-..............--.....(((....))) ( -9.50, z-score = -2.24, R) >droAna3.scaffold_13266 14957252 64 - 19884421 AACGAAAACCAGUUUAAUGAACCUUUAAGCUGAAAAAUAUUUCAGCUAGCAGCUCCGAGCUAGC .........((((((((.......))))))))............((((((........)))))) ( -15.40, z-score = -2.65, R) >dp4.chr3 4096069 60 - 19779522 AACGAAAACCAGUUUAAUGAACCUUUAAGCU--CAGGCAUUCCAAGC--GGGGCGCGGAUUGGG ........(((((((..(((.(......).)--)).((.((((....--)))).))))))))). ( -11.70, z-score = 0.70, R) >droPer1.super_2 4283147 60 - 9036312 AACGAAAACCAGUUUAAUGAACCUUUAAGCU--CAGGCAUUCCAAGC--GGGGCGCGGAUUGGG ........(((((((..(((.(......).)--)).((.((((....--)))).))))))))). ( -11.70, z-score = 0.70, R) >droVir3.scaffold_12875 13837890 59 - 20611582 AACUAAAACCAGUUUAAUGAACCUUUUAGCUGAAC-AUAUUUUCAAU--GGGACU--AGACAGC ..(((...(((((((...))))........((((.-.....)))).)--))...)--))..... ( -4.60, z-score = 0.77, R) >droMoj3.scaffold_6496 9250797 59 - 26866924 AACUAAAACCAGUUUAAUGAACCUUUAAGCUGAAA-AUAUUUUCAAU--GGGACU--AGACAGC ..(((...(((((((((.......))))))(((((-....))))).)--))...)--))..... ( -9.00, z-score = -1.29, R) >droGri2.scaffold_15245 927808 62 + 18325388 AACUAAAACCAGUUUAAUGAACUUUUAAGCUGAACAAUAUGUUCAAU--GGGUCUAAAGACAGC ........(((((((((.......))))))(((((.....))))).)--))(((....)))... ( -12.60, z-score = -2.07, R) >consensus AACAAAAACCAGUUUAAUGAACCUUUAAGCUG_AAAAUAUUUCGACU__CGGCCGCGAACUAGC .........((((((((.......))))))))................................ ( -5.30 = -5.57 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:03 2011