| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,573,984 – 18,574,078 |
| Length | 94 |
| Max. P | 0.992715 |

| Location | 18,573,984 – 18,574,078 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.56 |
| Shannon entropy | 0.28538 |
| G+C content | 0.37334 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.50 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
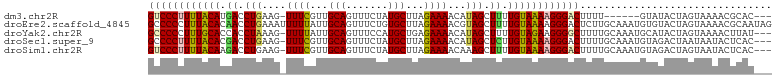
>dm3.chr2R 18573984 94 + 21146708 ---GUGCGUUUUACUAGUAUAC------AAAAGUCCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAA-CUUCAGGUCAUGUAAAAGGGAC ---((((.........))))..------....((((((((((((....(((((((.....)))))))...(((.((.....-.)))))....)))))))))))) ( -24.70, z-score = -3.09, R) >droEre2.scaffold_4845 19934116 104 + 22589142 CUAUUGCGUUUUACUAGUACACAUUUGCAAGAGUCCCUUUUACAAAAGCUACGUUUUCUAAGCACAGAAACUGCAAUAAAAAUUUCAGGUUGUGUAAAGGGGGC ...(((((.................)))))..(((((((((((....(((..........)))((((...(((.(((....))).))).))))))))))))))) ( -23.93, z-score = -1.16, R) >droYak2.chr2R 16039422 100 + 21139217 ---AUAAGUUUUACUAGUAUGCAUUUGCAAAAGCCCCUUCUACAAAAGCUAUGUUUUCUCAGCAUGGAAACUGCAAUAAAA-CUUUAGGUGGUGCAAAGGGGGC ---................(((....)))...(((((((...((..(.(((.(((((....(((.(....))))...))))-)..))).)..))...))))))) ( -24.90, z-score = -0.55, R) >droSec1.super_9 1876263 100 + 3197100 ---GUGAGUAUUAUUAGUCUACAUUUGCAAAAGUCCCUUUUACAAGAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAA-CUUCAGGUCGUGUAAAAGGGGC ---((((.((....)).)).))..........((((((((((((.((.((..(((((....(((.......)))...))))-)...)).)).)))))))))))) ( -26.90, z-score = -2.51, R) >droSim1.chr2R 17167970 100 + 19596830 ---GUGAGUAUUACUAGUCUACAUUUGCAAAAGUCCCUUUUACAAAAGCUUUGUUUUCUAAGCAUAGAAACUGCAACGAAA-CUUCAGGUCUUGUAAAAGGGAC ---(..(((.............)))..)....(((((((((((((.(.((..(((((....(((.......)))...))))-)...)).).))))))))))))) ( -24.72, z-score = -2.11, R) >consensus ___GUGAGUUUUACUAGUAUACAUUUGCAAAAGUCCCUUUUACAAAAGCUAUGUUUUCUAAGCAUAGAAACUGCAACGAAA_CUUCAGGUCGUGUAAAAGGGGC ................................((((((((((((....(((((((.....)))))))...(((.((.......)))))....)))))))))))) (-20.46 = -20.50 + 0.04)
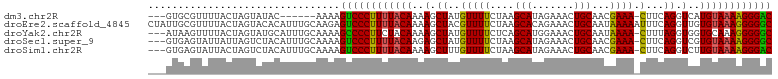
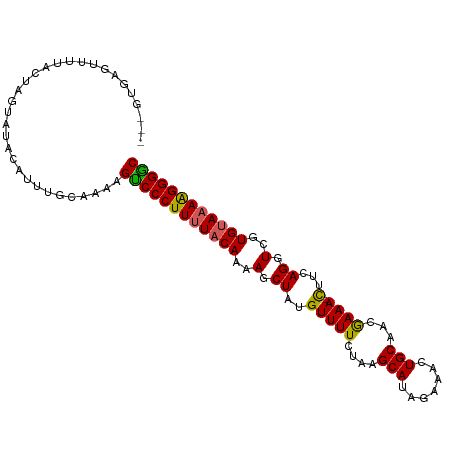
| Location | 18,573,984 – 18,574,078 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Shannon entropy | 0.28538 |
| G+C content | 0.37334 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.66 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.992715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
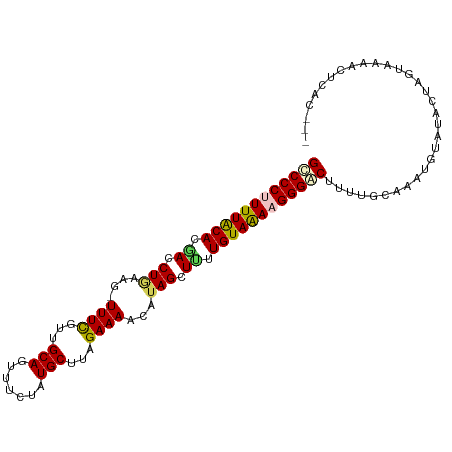
>dm3.chr2R 18573984 94 - 21146708 GUCCCUUUUACAUGACCUGAAG-UUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUUUUGUAAAAGGGACUUUU------GUAUACUAGUAAAACGCAC--- ((((((((((((.((.(((...-((((...(((.......)))...))))...))).)).))))))))))))....------...................--- ( -26.80, z-score = -3.51, R) >droEre2.scaffold_4845 19934116 104 - 22589142 GCCCCCUUUACACAACCUGAAAUUUUUAUUGCAGUUUCUGUGCUUAGAAAACGUAGCUUUUGUAAAAGGGACUCUUGCAAAUGUGUACUAGUAAAACGCAAUAG (((((.((((((.((.(((...((((((..((((...))))...))))))...))).)).)))))).)))(((..((((....))))..))).....))..... ( -20.80, z-score = -0.73, R) >droYak2.chr2R 16039422 100 - 21139217 GCCCCCUUUGCACCACCUAAAG-UUUUAUUGCAGUUUCCAUGCUGAGAAAACAUAGCUUUUGUAGAAGGGGCUUUUGCAAAUGCAUACUAGUAAAACUUAU--- (((((.((((((....(((..(-((((....((((......))))..))))).)))....)))))).)))))...(((....)))................--- ( -25.60, z-score = -1.78, R) >droSec1.super_9 1876263 100 - 3197100 GCCCCUUUUACACGACCUGAAG-UUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUCUUGUAAAAGGGACUUUUGCAAAUGUAGACUAAUAAUACUCAC--- ..((((((((((.((.(((...-((((...(((.......)))...))))...))).)).))))))))))...(((((....)))))..............--- ( -25.90, z-score = -3.09, R) >droSim1.chr2R 17167970 100 - 19596830 GUCCCUUUUACAAGACCUGAAG-UUUCGUUGCAGUUUCUAUGCUUAGAAAACAAAGCUUUUGUAAAAGGGACUUUUGCAAAUGUAGACUAGUAAUACUCAC--- (((((((((((((((.((....-((((...(((.......)))...))))....)).))))))))))))))).(((((....)))))..............--- ( -31.60, z-score = -4.32, R) >consensus GCCCCUUUUACACGACCUGAAG_UUUCGUUGCAGUUUCUAUGCUUAGAAAACAUAGCUUUUGUAAAAGGGACUUUUGCAAAUGUAUACUAGUAAAACUCAC___ ((((((((((((.((.(((....((((...(((.......)))...))))...))).)).))))))))))))................................ (-21.54 = -21.66 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:44:00 2011