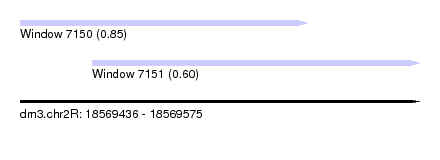
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,569,436 – 18,569,575 |
| Length | 139 |
| Max. P | 0.850225 |

| Location | 18,569,436 – 18,569,536 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 66.68 |
| Shannon entropy | 0.56449 |
| G+C content | 0.54039 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -16.53 |
| Energy contribution | -17.51 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
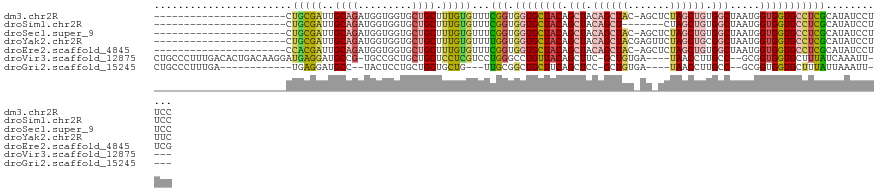
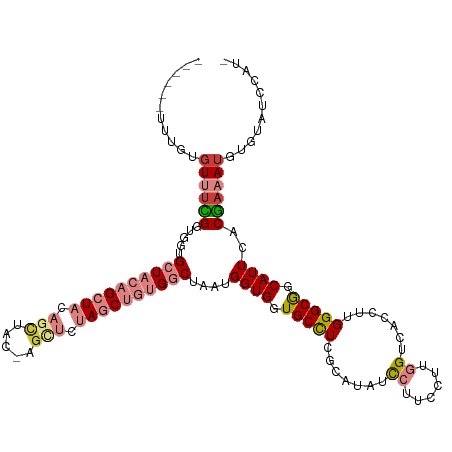
>dm3.chr2R 18569436 100 + 21146708 ----------------------CUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCC ----------------------.(((((..((.....((((..(..((.........))..)..)))).(((((((((((-(((....)))))))))....))))))).)))))......... ( -38.80, z-score = -1.56, R) >droSim1.chr2R 17163382 94 + 19596830 ----------------------CUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCU-------CUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCC ----------------------.(((((..(((....((((..(..((.........))..)..))))(((((((((.-------...)))))))))........))).)))))......... ( -36.00, z-score = -1.76, R) >droSec1.super_9 1871671 100 + 3197100 ----------------------CUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCC ----------------------.(((((..((.....((((..(..((.........))..)..)))).(((((((((((-(((....)))))))))....))))))).)))))......... ( -38.80, z-score = -1.56, R) >droYak2.chr2R 16034595 101 + 21139217 ----------------------CUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUUGGUGGUGCUACAGCUACAGCUACGAGUUCUAGCUGCGGCUAAUGGUGGUGCCUCGCAUAUCCUUUC ----------------------.(((((..(((....((((..(..((.........))..)..))))(((..((((((.(....)))))))..)))........))).)))))......... ( -35.10, z-score = -0.76, R) >droEre2.scaffold_4845 19929401 100 + 22589142 ----------------------CCACGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCG ----------------------.(((((..((((.........)))).)))))...(((.((..(((((((((((((((.-.....)))))))))))....))))..)))))........... ( -37.10, z-score = -1.17, R) >droVir3.scaffold_12875 13779470 111 + 20611582 CUGCCCUUUGACACUGACAAGGAUGAGGAUGCCG-UGCCGCUGCUGCUCCUCGUCCUGGGCCUGUUACAGCUUC-GCUGUGA----UAACCUUGCC--GCGGUGGUGCUUUAUCAAAUU---- ...........(((((.(.((((((((((.((.(-(......)).)))))))))))).(((.(((((((((...-)))))))----)).....)))--))))))...............---- ( -39.50, z-score = -2.21, R) >droGri2.scaffold_15245 861771 95 - 18325388 CUGCCCUUUGA------------UGAGGAUGCC--UACUCCUGCUGCUGCUG---UUGCGGCUGCUUCAGCUCC-GCUGUGA----UAACCUUGCC--GCGGUGGUGCUUUAUUAAAUU---- ......(((((------------((((((....--...))).((.(((((..---..))))).))...(((.((-((((((.----.........)--))))))).)))))))))))..---- ( -30.90, z-score = -2.33, R) >consensus ______________________CUGCGAUUGCAGAUGGUGGUGCUGCUUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC_AG_UCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCC .......................(((((..((((.........)))).)))))...(((.((((((((((((.((((((.......)))))).))))....)))))))))))........... (-16.53 = -17.51 + 0.99)
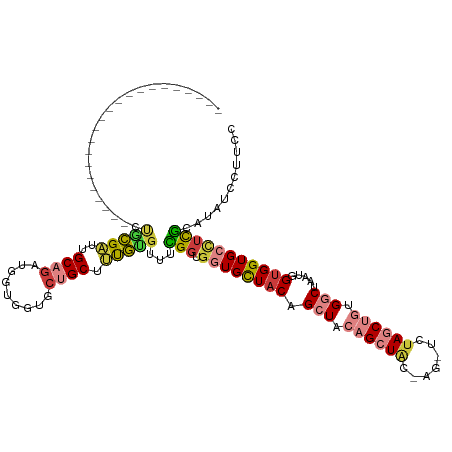
| Location | 18,569,461 – 18,569,575 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Shannon entropy | 0.42433 |
| G+C content | 0.50704 |
| Mean single sequence MFE | -37.11 |
| Consensus MFE | -23.00 |
| Energy contribution | -25.04 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18569461 114 + 21146708 ------UUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCAUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU- ------...(..((((((.((..(((((((((((((((.-.....)))))))))))....))))..)).)......((.((((.((...)).)))).))......)))))..)........- ( -40.40, z-score = -1.67, R) >droSim1.chr2R 17163407 108 + 19596830 ------UUUGUGUUUCGGUGGUGCUACAGCUACAG-------CUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU- ------...(..((((((.((..((((((((((((-------(....)))))))))....))))..)).)......((.(((..((...))..))).))......)))))..)........- ( -37.70, z-score = -1.81, R) >droSec1.super_9 1871696 114 + 3197100 ------UUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU- ------...(..(((((..(((((((((((((.(((...-.))).))))))).(((...((((((((...))))..((......)).))))...)))))))))..)))))..)........- ( -40.30, z-score = -1.81, R) >droYak2.chr2R 16034620 115 + 21139217 ------UUUGUGUUUUGGUGGUGCUACAGCUACAGCUACGAGUUCUAGCUGCGGCUAAUGGUGGUGCCUCGCAUAUCCUUUCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU- ------...(..(((((..(((((..((((((.(((.....))).))))))((.((((.((((((((...))))..((......)).)))))))).)))))))..)))))..)........- ( -34.70, z-score = -0.18, R) >droEre2.scaffold_4845 19929426 114 + 22589142 ------UUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC-AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCGUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU- ------..........((.((..(((((((((((((((.-.....)))))))))))....))))..))..((((((..(((((..(((.((...)).)))....)))))))))))..))..- ( -40.70, z-score = -1.87, R) >droVir3.scaffold_12875 13779510 111 + 20611582 UGCUGCUCCUCGUCCUGGGCCUGUUACAGCUUCGCU-----GUGAUAACCUUGCC--GCGGUGGUGCUU----UAUCAAAUUCUGGUCACCUUGGGCGGCAUUUACAAAAUGUGUAUCCACU ((((((((.((((...(((..(((((((((...)))-----)))))).)))....--)))).((((((.----...........)).))))..))))))))..........(((....))). ( -32.70, z-score = -0.88, R) >droGri2.scaffold_15245 861795 111 - 18325388 UCCUGCUGCUGCUGUUGCGGCUGCUUCAGCUCCGCU-----GUGAUAACCUUGCC--GCGGUGGUGCUU----UAUUAAAUUCUGGUCACCUUGGGCAGCAUUUACAAAAUGUGUAUCCACU ....((.(((((....))))).))...(((.(((((-----(((..........)--))))))).))).----...(((((.(((.((.....)).))).)))))......(((....))). ( -33.30, z-score = -0.73, R) >consensus ______UUUGUGUUUCGGUGGUGCUACAGCUACAGCUAC_AGCUCUAGCUGUGGCUAAUGGUGGUGCCUCGCAUAUCCUUCCUUGGUCACCUUGGGCGGCAUUCACGAAAUGUGUAUCCAU_ ...........((((((.....((((((((((.(((.....))).))))))))))....((((.(((((.......((......)).......))))).))))..))))))........... (-23.00 = -25.04 + 2.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:58 2011