| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,484,791 – 18,484,900 |
| Length | 109 |
| Max. P | 0.615052 |

| Location | 18,484,791 – 18,484,900 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.19 |
| Shannon entropy | 0.71128 |
| G+C content | 0.38005 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -6.95 |
| Energy contribution | -7.74 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.615052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
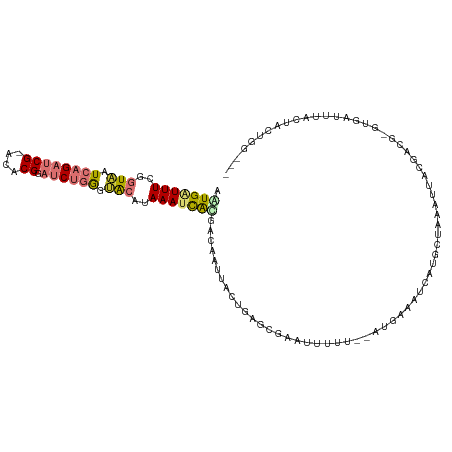
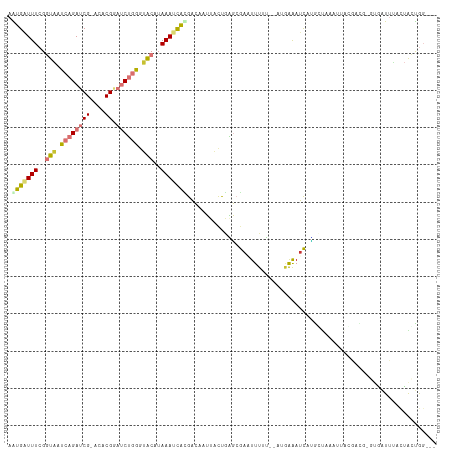
>dm3.chr2R 18484791 109 - 21146708 AAUGAUUUCGGUAAUCAGAUCG-ACACGGAUCUGGGUACAUAAAUCGCGACACUUACUGAGCGAUUUUUU--AUGAAAUCAUGCUUUAUUACGACGUGUGAUUUACUACUGG--- .((((((((((((..((((((.-.....))))))..)))..(((((((............)))))))...--.))))))))).....(((((.....)))))..........--- ( -27.70, z-score = -2.65, R) >droSec1.super_9 1783436 109 - 3197100 AAUGAUUUCGGUAAUCAGAUCG-ACACGGAUCUGGGUACAUAAAUCGCGACAAUUACUGGGCGAUUUUUU--AUGAAAUCAUGGUGUAUUACGAUGGGUGAUUUACUACUGG--- .((((((((((((..((((((.-.....))))))..)))..(((((((..((.....)).)))))))...--.)))))))))((((.(((((.....))))).)))).....--- ( -33.10, z-score = -4.02, R) >droYak2.chr2R 15947306 101 - 21139217 AAUGAUUUCGGUAAUCAGAUCG-ACACGGAUCUGGGUACUUAAAGCAUGACAAUUACCGAGCGAAUUUUUUGAAGAAAUCAUGCCAAU----------UAUUUGACUACUGG--- .((((((((((((..((((((.-.....))))))..))))....((.((........)).))............))))))))......----------..............--- ( -22.10, z-score = -1.55, R) >droEre2.scaffold_4845 15117218 98 + 22589142 AGUGAUUUCGGUAAUCAGAUCG-ACACGGAUCUGGGUACAUAAAGCGUGACAAUUACUGAGCGAAUUUGU--ACGAAAUCAUGCUAAA-----------GAUUUACUACUGG--- .((((((((.....(((((((.-.....)))))))(((((.....(((..((.....)).)))....)))--))))))))))......-----------.............--- ( -25.30, z-score = -2.22, R) >droAna3.scaffold_13266 12610848 113 + 19884421 AGUGAUUUCGGUAAUCAGAUCG-ACACGAAUCUGAAAACAUAAAUUACGUCCCUUACUUGGCACUUUUUUUAUUCGAAAUUCAUUCAUCCAAGAUGUGAUAUUUGAUGGUAUAG- .(((((((..((..((((((((-...)).))))))..))..)))))))((.((......)).)).........(((((..((((((......)).))))..)))))........- ( -19.70, z-score = 0.12, R) >dp4.chr3 7091057 113 - 19779522 AGUGGUUUCGUUUAUCAGAUCGCGCACGUUCCAUGGAGCAUAAAUCAUGAAACAUUUCAUUAAAAUUUGG--AAAAGAUUCAGUCUCGUUCACAUGAUUCGUGAAUCGACACUAU ((((...((........))(((..(((((((....))))...(((((((.(((.(((((........)))--)).((((...)))).)))..))))))).)))...))))))).. ( -22.20, z-score = 0.06, R) >droPer1.super_2 7290598 112 - 9036312 AGUGGUUUCGUUUAUCAGAUCGCGCACGUUCCAUGGAGCAUAAAUCAUGAAACAUUUCAUUAAAAUUUGG--AAAAGAUUCGGUC-AAUCGGCACUAUUGAUGGAACGAUGGAAU .(((((((........))))))).((((((((((.((..........((((...(((((........)))--))....))))(((-....)))....)).)))))))).)).... ( -23.30, z-score = 0.13, R) >droSim1.chr2R 17069331 109 - 19596830 AAUGAUUUCGGUAAUCAGAUCG-ACACGGAUCUGGGUACAUAAAUCGCGACAAUUACUGAGCGAUUUUUU--AUGAAAUCAUGGUGUAUUACGACGGGUGAUUUACUACUGG--- .((((((((((((..((((((.-.....))))))..)))..(((((((..((.....)).)))))))...--.)))))))))((((.(((((.....))))).)))).....--- ( -32.70, z-score = -3.77, R) >consensus AAUGAUUUCGGUAAUCAGAUCG_ACACGGAUCUGGGUACAUAAAUCACGACAAUUACUGAGCGAAUUUUU__AUGAAAUCAUGCUAAAUUACGACG_GUGAUUUACUACUGG___ .(((((((..(((.((((((((....)).)))))).)))..)))))))................................................................... ( -6.95 = -7.74 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:48 2011