
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,481,547 – 18,481,646 |
| Length | 99 |
| Max. P | 0.509423 |

| Location | 18,481,547 – 18,481,646 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.31 |
| Shannon entropy | 0.73621 |
| G+C content | 0.52795 |
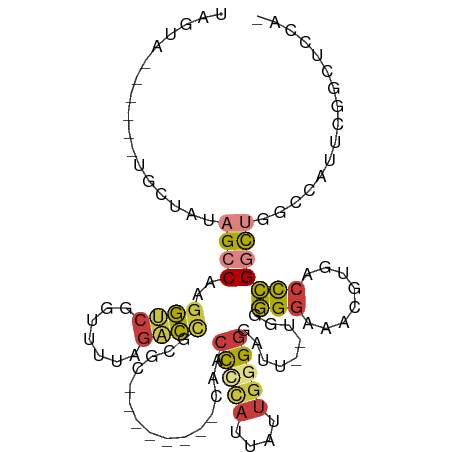
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.49 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.38 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
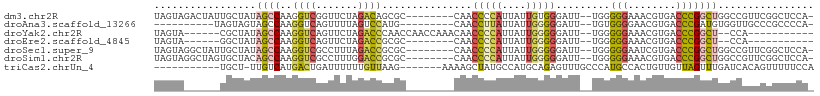
>dm3.chr2R 18481547 99 - 21146708 UAGUAGACUAUUGCUAUAGCCAAGGUCGGUUCUAGACAGCGC--------CAACCCCAUUAUUGUGGGAUU--UGGGGGAAACGUGACCCGGCUGGCCGUUCGGCUCCA- ((((((....)))))).((((..((((((((..........(--------(((.(((((....)))))..)--)))(((........)))))))))))....))))...- ( -31.50, z-score = 0.16, R) >droAna3.scaffold_13266 12607397 88 + 19884421 ----------UAGUAGUAGCCAAGGUCAGUUUUAGUCCAUG---------CAACCUUAUUAUUGGGGGAUU--UGUGGGGAACGUGACCCGAUGUGGUUGCCCGCCCCA- ----------.....(((((((.(((((((((...(((((.---------...(((((....)))))....--.))))))))).))))).....)))))))........- ( -26.10, z-score = -0.11, R) >droYak2.chr2R 15945666 89 - 21139217 UAGUA------CGCUAUAGCCAAGGUCAGUUCUAGACCCAACCAACCAAACAACCCCAUUAUUGGGGGAUU--UGGGGGAAACGUGACCCGGCU--CCA----------- (((..------..))).((((..((((.......)))).......(((((...(((((....)))))..))--)))(((........)))))))--...----------- ( -28.60, z-score = -1.14, R) >droEre2.scaffold_4845 15115757 81 + 22589142 UAGUA------GGCUAUAGCCAAGGUCAGUUCUAGACCGCGC--------CAACCCCAUUAUUGGGGGAUU--UGGGGGAAACGUGACCCGGCU--CCA----------- ..((.------(((....)))..((((.......))))))((--------(..(((((....)))))....--.((((....)....)))))).--...----------- ( -29.80, z-score = -1.04, R) >droSec1.super_9 1780140 99 - 3197100 UAGUAGGCUAUUGCUAUAGCCAAGGUCGCCUUUAGACCGCGC--------CAACCCCAUUAUUGGGGGAUU--UGGGGGAAUCGUGACCCGGCUGGCCGUUCGGCUCCA- ..((.((((((....))))))..((((.......))))))((--------(..(((((....)))))....--..(((.........)))))).((((....))))...- ( -38.90, z-score = -0.79, R) >droSim1.chr2R 17065662 99 - 19596830 UAGUAGGCUAGUGCUACAGCCAAGGUCGCCUUUGGACCGCGC--------CAACCCCAUUAUUGGGGGAUU--UGGGGGAAACGUGACCCGGCUGGCCGUUCGGCUCCA- .....(((....))).(((((..((((((.(((...((.((.--------...(((((....)))))....--)).)).))).)))))).)))))(((....)))....- ( -39.30, z-score = -0.31, R) >triCas2.chrUn_4 920648 91 + 1175385 -----------UGCU-UUGUCAUGACUGAUUUUUUGUUAAG-------AAAAGCUAUGCCAUGCAGAGUUUGCCCAUGCCACUGUUGUUAGUUUGAUCACAGUUUUUCCA -----------.(((-(((.((((.(...((((((....))-------)))).....).))))))))))...........(((((.(((.....))).)))))....... ( -15.50, z-score = 0.55, R) >consensus UAGUA______UGCUAUAGCCAAGGUCGGUUUUAGACCGCGC________CAACCCCAUUAUUGGGGGAUU__UGGGGGAAACGUGACCCGGCUGGCCAUUCGGCUCCA_ .................((((..((((.......))))...............(((((....))))).........(((........)))))))................ (-14.05 = -14.49 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:48 2011