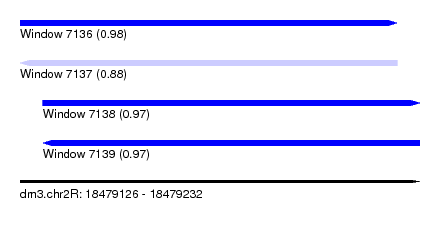
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,479,126 – 18,479,232 |
| Length | 106 |
| Max. P | 0.978207 |

| Location | 18,479,126 – 18,479,226 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 67.64 |
| Shannon entropy | 0.53571 |
| G+C content | 0.40897 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -11.38 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18479126 100 + 21146708 UUUAGUGCUACCUCGUAAUAUCCUCUUAUAGAGGAUACGUAAUCCAGUAGGGGUACAUCAUAUAUCACACA-CGUGUGAC---C--------AAAUUGCAUAAACUGCGGUC ....(((.((((((......(((((.....)))))(((........))))))))))))......(((((..-..))))).---.--------..((((((.....)))))). ( -25.30, z-score = -1.65, R) >droYak2.chr2R 15942998 112 + 21139217 UUUAUUGUCACACCGUAAUAUCCUCUUAUAGAGGAUACGUAAUUCAGUGGUUAUGUACAUCACAUCACACAUCGCACAACACACGUGUGUCGAAAUUGCGUAAACCACGUUU .............(((....(((((.....)))))(((((((((..(((...((((.....))))..))).(((((((.......)))).))))))))))))....)))... ( -26.50, z-score = -1.92, R) >droEre2.scaffold_4845 15113264 90 - 22589142 UUUGGUGUCACAUCGUAAUAUCCUCUUAUAGAGGAUACGUAA---------UUCAGUGGGUAUAUCACAAA-CGUGUGGC---A--------AAAUUGCGUAAACUACGUU- .....(((((((.(((..(((((((.....))))))).....---------....((((.....))))..)-))))))))---)--------.....(((((...))))).- ( -25.90, z-score = -3.22, R) >droSec1.super_9 1777623 81 + 3197100 UUUAGUGCUACCUCGUAAUAUCCUCCUUUAGAGG-UAC---------------------ACAUAUCACACA-CGUGUGACCACC--------AAAUUGCAUAAACUGCGGUC ....(((.((((((.(((.........)))))))-)))---------------------))...(((((..-..))))).....--------..((((((.....)))))). ( -19.50, z-score = -2.35, R) >droSim1.chr2R 17063159 81 + 19596830 UUUAGUGCUACCUCGUAAUAUCCUCUUAUAGGGG-UAC---------------------ACAUAUCACACA-CGUGUGACCACC--------AAAUUGCAUAAACUGCGGUC ....(((.((((((...(((......))).))))-)))---------------------))...(((((..-..))))).....--------..((((((.....)))))). ( -19.20, z-score = -1.55, R) >consensus UUUAGUGCUACCUCGUAAUAUCCUCUUAUAGAGGAUACGUAA_________________ACAUAUCACACA_CGUGUGAC___C________AAAUUGCAUAAACUGCGGUC .............((((.(((((((.....)))))))...........................(((((.....)))))..........................))))... (-11.38 = -12.22 + 0.84)

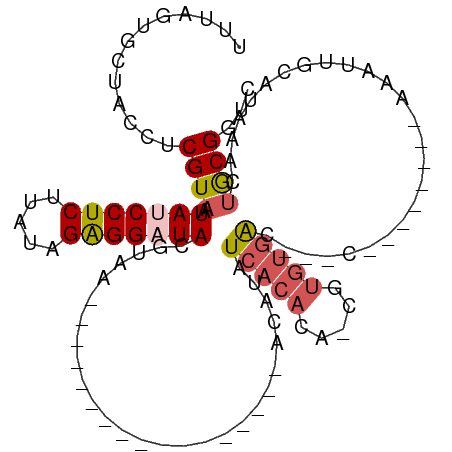
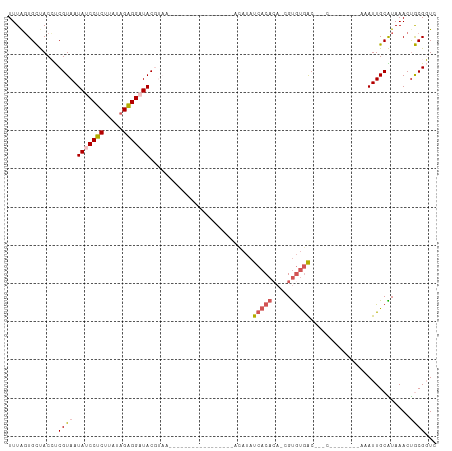
| Location | 18,479,126 – 18,479,226 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 67.64 |
| Shannon entropy | 0.53571 |
| G+C content | 0.40897 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18479126 100 - 21146708 GACCGCAGUUUAUGCAAUUU--------G---GUCACACG-UGUGUGAUAUAUGAUGUACCCCUACUGGAUUACGUAUCCUCUAUAAGAGGAUAUUACGAGGUAGCACUAAA .((((((.....)))...((--------(---((((((..-..)))))).......(((..((....))..)))((((((((.....))))))))..))))))......... ( -26.50, z-score = -1.46, R) >droYak2.chr2R 15942998 112 - 21139217 AAACGUGGUUUACGCAAUUUCGACACACGUGUGUUGUGCGAUGUGUGAUGUGAUGUACAUAACCACUGAAUUACGUAUCCUCUAUAAGAGGAUAUUACGGUGUGACAAUAAA ..((((((((((((((....((((((....))))))))))..(((.(.((((.....)))).))))))))))))))((((((.....))))))................... ( -31.40, z-score = -1.31, R) >droEre2.scaffold_4845 15113264 90 + 22589142 -AACGUAGUUUACGCAAUUU--------U---GCCACACG-UUUGUGAUAUACCCACUGAA---------UUACGUAUCCUCUAUAAGAGGAUAUUACGAUGUGACACCAAA -.((((((((((.(((....--------)---))((((..-..))))..........))))---------))))))((((((.....))))))................... ( -20.80, z-score = -2.67, R) >droSec1.super_9 1777623 81 - 3197100 GACCGCAGUUUAUGCAAUUU--------GGUGGUCACACG-UGUGUGAUAUGU---------------------GUA-CCUCUAAAGGAGGAUAUUACGAGGUAGCACUAAA ....(((.....)))..(((--------((((((((((..-..)))))).(((---------------------(((-((((.....)))).)).))))......))))))) ( -22.10, z-score = -1.12, R) >droSim1.chr2R 17063159 81 - 19596830 GACCGCAGUUUAUGCAAUUU--------GGUGGUCACACG-UGUGUGAUAUGU---------------------GUA-CCCCUAUAAGAGGAUAUUACGAGGUAGCACUAAA .((((((.....))).....--------((.(((.(((((-(((...))))))---------------------)))-))))..................)))......... ( -18.90, z-score = -0.03, R) >consensus GACCGCAGUUUAUGCAAUUU________G___GUCACACG_UGUGUGAUAUGU_________________UUACGUAUCCUCUAUAAGAGGAUAUUACGAGGUAGCACUAAA ....(((.....))).................(((((((...))))))).........................((((((((.....))))))))................. (-13.62 = -13.70 + 0.08)

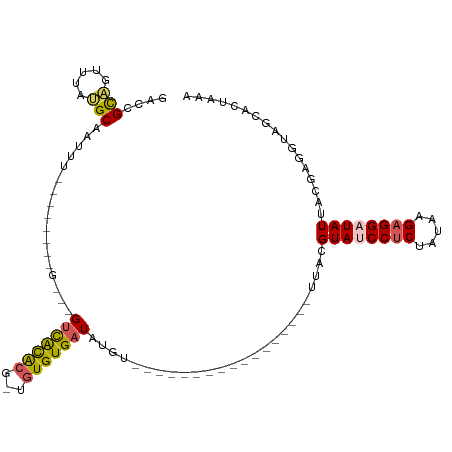
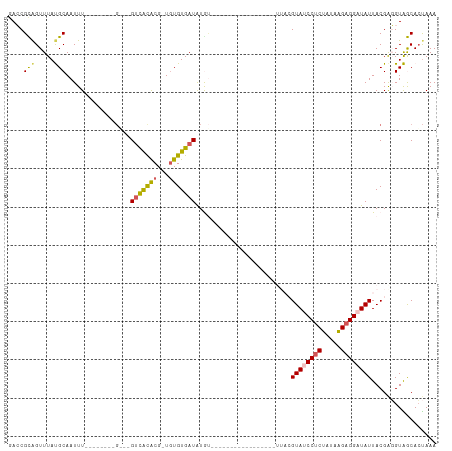
| Location | 18,479,132 – 18,479,232 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.51267 |
| G+C content | 0.42641 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -11.51 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18479132 100 + 21146708 GCUACCUCGUAAUAUCCUCUUAUAGAGGAUACGUAAUCCAGUAGGGGUACAUCAUAUAUCACAC------------ACGUGUGACC---AAAUUGCAUAAACUGCGGUCGAGGAU ....(((((...(((((((.....))))))).(((..((....))..)))........(((((.------------...)))))..---..((((((.....))))))))))).. ( -29.30, z-score = -2.53, R) >droYak2.chr2R 15943004 112 + 21139217 GUCACACCGUAAUAUCCUCUUAUAGAGGAUACGUAAUUCAGUGGUUAUGUACAUCACAUCACACAUCGCACAACACACGUGUGUCG---AAAUUGCGUAAACCACGUUUGAAGAU .(((...(((....(((((.....)))))(((((((((..(((...((((.....))))..))).(((((((.......)))).))---))))))))))....)))..))).... ( -27.60, z-score = -1.60, R) >droEre2.scaffold_4845 15113270 90 - 22589142 GUCACAUCGUAAUAUCCUCUUAUAGAGGAUACGUAAUUCAGUGG----GUAUAUCACAA-----------------ACGUGUGGCA---AAAUUGCGUAAACUACGUU-GAAGAU ((((((.(((..(((((((.....))))))).........((((----.....))))..-----------------))))))))).---.....(((((...))))).-...... ( -25.00, z-score = -3.05, R) >droSec1.super_9 1777629 81 + 3197100 GCUACCUCGUAAUAUCCUCCUUUAGAGG-UAC---------------------ACAUAUCACAC------------ACGUGUGACCACCAAAUUGCAUAAACUGCGGUCGAGGAU ....(((((......((((.....))))-...---------------------.....(((((.------------...))))).......((((((.....))))))))))).. ( -19.80, z-score = -1.98, R) >droSim1.chr2R 17063165 81 + 19596830 GCUACCUCGUAAUAUCCUCUUAUAGGGG-UAC---------------------ACAUAUCACAC------------ACGUGUGACCACCAAAUUGCAUAAACUGCGGUCGAGGAU ....(((((...............((((-(.(---------------------((((.......------------..)))))))).))..((((((.....))))))))))).. ( -21.80, z-score = -2.15, R) >consensus GCUACCUCGUAAUAUCCUCUUAUAGAGGAUACGUAAU_CAGU_G______A__ACAUAUCACAC____________ACGUGUGACC___AAAUUGCAUAAACUGCGGUCGAGGAU ....(((((...(((((((.....)))))))..............................................(((((..................)).)))..))))).. (-11.51 = -12.19 + 0.68)

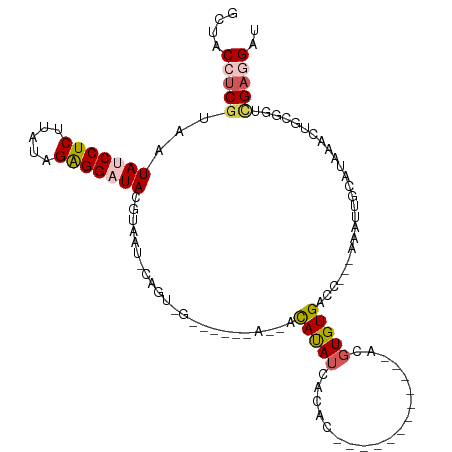
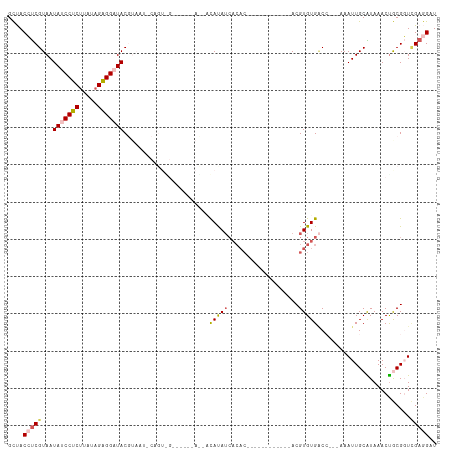
| Location | 18,479,132 – 18,479,232 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.66 |
| Shannon entropy | 0.51267 |
| G+C content | 0.42641 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -14.32 |
| Energy contribution | -16.68 |
| Covariance contribution | 2.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18479132 100 - 21146708 AUCCUCGACCGCAGUUUAUGCAAUUU---GGUCACACGU------------GUGUGAUAUAUGAUGUACCCCUACUGGAUUACGUAUCCUCUAUAAGAGGAUAUUACGAGGUAGC ..(((((...(((.....))).....---.((((((...------------.)))))).......(((..((....))..)))((((((((.....))))))))..))))).... ( -30.20, z-score = -2.32, R) >droYak2.chr2R 15943004 112 - 21139217 AUCUUCAAACGUGGUUUACGCAAUUU---CGACACACGUGUGUUGUGCGAUGUGUGAUGUGAUGUACAUAACCACUGAAUUACGUAUCCUCUAUAAGAGGAUAUUACGGUGUGAC ....(((.((((((((((((((....---((((((....)))))).....)))))))((((.....)))))))))........((((((((.....))))))))....)).))). ( -32.80, z-score = -1.44, R) >droEre2.scaffold_4845 15113270 90 + 22589142 AUCUUC-AACGUAGUUUACGCAAUUU---UGCCACACGU-----------------UUGUGAUAUAC----CCACUGAAUUACGUAUCCUCUAUAAGAGGAUAUUACGAUGUGAC (((...-.((((((((((.(((....---)))((((...-----------------.))))......----....))))))))))((((((.....)))))).....)))..... ( -21.40, z-score = -2.62, R) >droSec1.super_9 1777629 81 - 3197100 AUCCUCGACCGCAGUUUAUGCAAUUUGGUGGUCACACGU------------GUGUGAUAUGU---------------------GUA-CCUCUAAAGGAGGAUAUUACGAGGUAGC ((((((....(((.....)))..(((((.(((.((((((------------((...))))))---------------------)))-)).))))).))))))............. ( -25.90, z-score = -2.13, R) >droSim1.chr2R 17063165 81 - 19596830 AUCCUCGACCGCAGUUUAUGCAAUUUGGUGGUCACACGU------------GUGUGAUAUGU---------------------GUA-CCCCUAUAAGAGGAUAUUACGAGGUAGC ((((((....(((.....))).....((.(((.((((((------------((...))))))---------------------)))-)))).....))))))............. ( -24.30, z-score = -1.71, R) >consensus AUCCUCGACCGCAGUUUAUGCAAUUU___GGUCACACGU____________GUGUGAUAUGU__U______C_ACUG_AUUACGUAUCCUCUAUAAGAGGAUAUUACGAGGUAGC ..(((((...(((.....))).........(((((((..............))))))).........................((((((((.....))))))))..))))).... (-14.32 = -16.68 + 2.36)

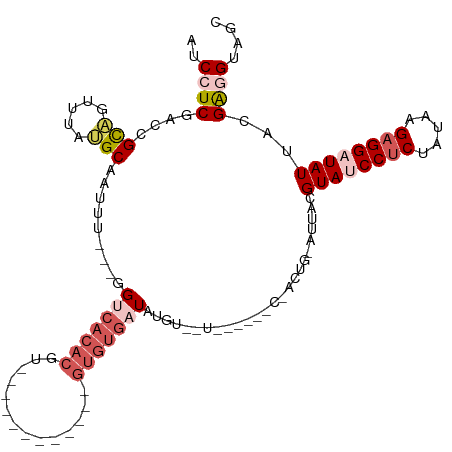
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:47 2011