| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,666,272 – 3,666,410 |
| Length | 138 |
| Max. P | 0.916181 |

| Location | 3,666,272 – 3,666,410 |
|---|---|
| Length | 138 |
| Sequences | 9 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Shannon entropy | 0.50905 |
| G+C content | 0.41034 |
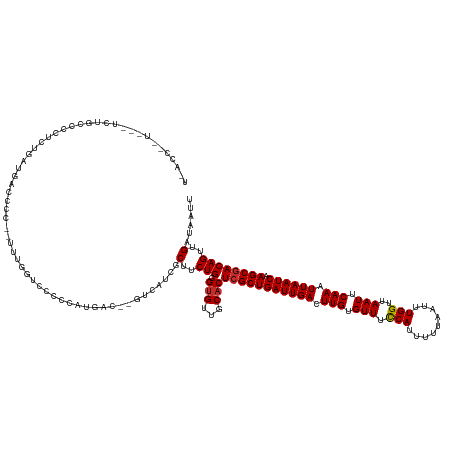
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -16.91 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
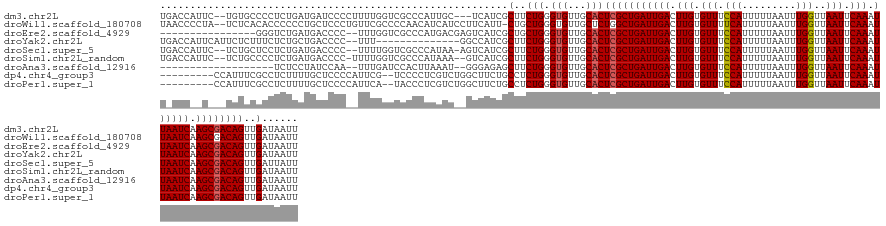
>dm3.chr2L 3666272 138 + 23011544 UGACCAUUC--UGUGCCCCUCUGAUGAUCCCCUUUUGGUCGCCCAUUGC---UCAUCGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU .(((.....--.((((..............((....)).((((((..((---.....))...))))))..))))(((((((((((.(((.(((.(((.........)))..))).))).)))))).)))))..)))....... ( -32.20, z-score = -1.54, R) >droWil1.scaffold_180708 9471119 140 + 12563649 UAACCCCUA--UCUCACACCCCCCUGCUCCCUGUUCGCCCCAACAUCAUCCUUCAUU-CUGCUGGGUGUUGCUCUGGCUGAUUGACUUGUGUUUUCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU .........--...................((((.(((...((((.((....(((.(-(.((..((......))..)).)).)))..)))))).......(((((((((.....)))))))))....)))))))......... ( -24.70, z-score = -0.53, R) >droEre2.scaffold_4929 3700514 125 + 26641161 ----------------GGGUCUGAUGACCCC--UUUGGUCGCCCAUGACGAGUCAUCGCUGCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU ----------------(((((....))))).--....(((((.....((((((((...(.((.(((((...))))))).)..))))))))..........(((((((((.....)))))))))....)))))........... ( -39.20, z-score = -2.81, R) >droYak2.chr2L 3659359 127 + 22324452 UGACCAUUCAUUCUCUUUCUCUGCUGACCCC--UUU--------------GGCCAUCGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU ...................((.(((((((((--...--------------(((....)))...))).)).....(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))).))..... ( -29.00, z-score = -1.82, R) >droSec1.super_5 1761925 138 + 5866729 UGACCAUUC--UCUGCUCCUCUGAUGACCCC--UUUUGGUCGCCCAUAA-AGUCAUCGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUUAUU .........--.....((..(((..((((..--....))))(((((..(-(((....)))).))))).......(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))..))..... ( -31.90, z-score = -1.62, R) >droSim1.chr2L_random 162550 138 + 909653 UGACCAUUC--UCUGCCCCUCUGAUGACCCC-UUUUGGUCGCCCAUAAA--GUCAUCGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU .....(((.--.(((.(((..(((((((...-...(((....)))....--))))))).....)))........(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))..))).... ( -31.30, z-score = -1.42, R) >droAna3.scaffold_12916 5721210 120 + 16180835 -------------------UCUCCUAUCCAA--UUUGAUCCACUUAAAU--GGGAGAGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU -------------------((((((((....--..............))--))))))....(((.(((...)))(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))......... ( -25.67, z-score = -0.65, R) >dp4.chr4_group3 5062469 132 + 11692001 ---------CCAUUUCGCCUCUUUUGCUCCCCAUUCG--UCCCCUCGUCUGGCUUCUGCCUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU ---------.....(((.((....(((.(((((..((--......))...(((....)))..)))).)..))).(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))).)).)))..... ( -28.20, z-score = -1.73, R) >droPer1.super_1 6554036 132 + 10282868 ---------CCAUUUCGCCUCUUUUGCUCCCCAUUCA--UACCCUCGUCUGGCUUCUGCCUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU ---------..........((..(((..........(--(((((......(((....)))...)))))).....(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))..))..... ( -29.20, z-score = -2.17, R) >consensus U_ACC__U___UCUGCCCCUCUGAUGACCCC__UUUGGUCCCCCAUGAC__GUCAUCGCUUCUGGGUGUUGCACUCGCUGAUUGACUUGUGUUUCCAUUUUUAAUUUGGUUAAUUCAAAUUAAUCAAGCGACAGUUGAUAAUU ..........................................................(..(((.(((...)))(((((((((((.(((.(((.(((.........)))..))).))).)))))).))))))))..)...... (-16.91 = -17.03 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:23 2011