
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,474,743 – 18,474,793 |
| Length | 50 |
| Max. P | 0.592994 |

| Location | 18,474,743 – 18,474,793 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 85.74 |
| Shannon entropy | 0.24851 |
| G+C content | 0.45081 |
| Mean single sequence MFE | -14.78 |
| Consensus MFE | -10.56 |
| Energy contribution | -11.28 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.592994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
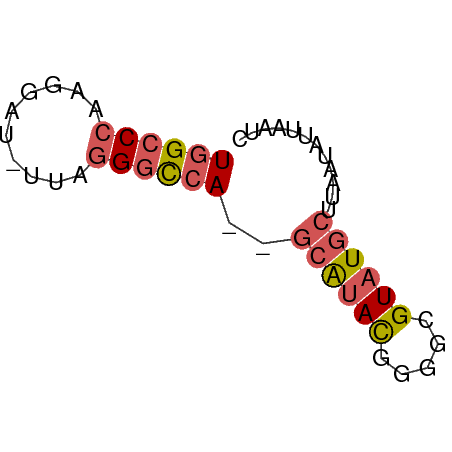
>dm3.chr2R 18474743 50 + 21146708 UGGCCCAAGGAU-UAAGGGCCA--GCAUAUGGGGCGUAUGCUUAAUAUUAAUC ((((((......-...))))))--(((((((...)))))))............ ( -17.20, z-score = -2.26, R) >droAna3.scaffold_13266 12601219 51 - 19884421 UAGGCCAAGGCUAUGAGGACCA--UAAAACAGGGCGUAUGCUUAAUAUUAAUC ((.(((..(..((((.....))--))...)..))).))............... ( -7.30, z-score = 0.43, R) >droYak2.chr2R 15938289 52 + 21139217 UGGCCCAAGGAU-UUAAGGCCAUGGCGUACAUGGCGUAUGCUUAAUAUUAAUC (((((.......-....))))).(((((((.....)))))))........... ( -14.10, z-score = -1.03, R) >droSec1.super_9 1773415 50 + 3197100 UGGCCCAAGGAU-UUAGGGUCA--GCAUACGGGGCGUAUGCUUAAUAUUAAUC ((((((......-...))))))--(((((((...)))))))............ ( -16.30, z-score = -2.94, R) >droSim1.chr2R 17058936 50 + 19596830 UGGCCCAAGGAU-UUAGGGCCA--GCAUACGGGGCGUAUGCUUAAUAUUAAUC ((((((......-...))))))--(((((((...)))))))............ ( -19.00, z-score = -3.13, R) >consensus UGGCCCAAGGAU_UUAGGGCCA__GCAUACGGGGCGUAUGCUUAAUAUUAAUC ((((((..........))))))..((((((.....))))))............ (-10.56 = -11.28 + 0.72)


| Location | 18,474,743 – 18,474,793 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 53 |
| Reading direction | reverse |
| Mean pairwise identity | 85.74 |
| Shannon entropy | 0.24851 |
| G+C content | 0.45081 |
| Mean single sequence MFE | -12.78 |
| Consensus MFE | -9.28 |
| Energy contribution | -10.12 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18474743 50 - 21146708 GAUUAAUAUUAAGCAUACGCCCCAUAUGC--UGGCCCUUA-AUCCUUGGGCCA ...........((((((.......)))))--)(((((...-......))))). ( -15.30, z-score = -2.35, R) >droAna3.scaffold_13266 12601219 51 + 19884421 GAUUAAUAUUAAGCAUACGCCCUGUUUUA--UGGUCCUCAUAGCCUUGGCCUA ..................(((..(..(((--((.....))))).)..)))... ( -5.00, z-score = 1.25, R) >droYak2.chr2R 15938289 52 - 21139217 GAUUAAUAUUAAGCAUACGCCAUGUACGCCAUGGCCUUAA-AUCCUUGGGCCA ............((.(((.....))).))..((((((...-......)))))) ( -10.70, z-score = -0.32, R) >droSec1.super_9 1773415 50 - 3197100 GAUUAAUAUUAAGCAUACGCCCCGUAUGC--UGACCCUAA-AUCCUUGGGCCA ...........((((((((...)))))))--)(.(((...-......))).). ( -12.90, z-score = -2.73, R) >droSim1.chr2R 17058936 50 - 19596830 GAUUAAUAUUAAGCAUACGCCCCGUAUGC--UGGCCCUAA-AUCCUUGGGCCA ...........((((((((...)))))))--)(((((...-......))))). ( -20.00, z-score = -4.27, R) >consensus GAUUAAUAUUAAGCAUACGCCCCGUAUGC__UGGCCCUAA_AUCCUUGGGCCA ............(((((((...)))))))...(((((..........))))). ( -9.28 = -10.12 + 0.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:43 2011