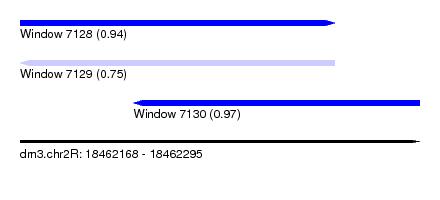
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,462,168 – 18,462,295 |
| Length | 127 |
| Max. P | 0.967724 |

| Location | 18,462,168 – 18,462,268 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.66753 |
| G+C content | 0.44433 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.99 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
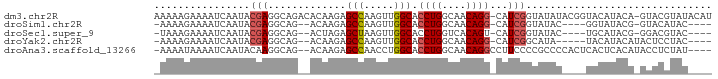
>dm3.chr2R 18462168 100 + 21146708 UUUAUAUAUAUUGUAGGUUGGUUGGUAUACGUACAUAUGUAUACGUACUGUAUGUACCGUAUAUACCGAUGCCUGUUGCCAGGUGCCAACUUGGCUCUUG .............(((((..(((((((((.(((((((((((....))).))))))))....))))))))))))))..(((((((....)))))))..... ( -33.10, z-score = -2.07, R) >droSim1.chr2R 17052496 79 + 19596830 ---------UUUAUAUAUAGGUUGGUGUACGUACGUA------UGUACCGUAUACC----GUAUACCGAUGCCUGUUGCCAGGUGCCAACUUGGCUCU-- ---------.......(((((((((((((((((((..------.....))))...)----))))))))..)))))).(((((((....)))))))...-- ( -29.50, z-score = -2.80, R) >droSec1.super_9 1767054 79 + 3197100 ---------AUAAUAUAUAGGAUGGUGUACGUACGUA------CGUCCCGUAUGCA----GUAUACCGAUGACUGUGACCAGGUGCCAACUUAGCUCU-- ---------......(((((..((((((((...((((------((...))))))..----))))))))....)))))...((((....))))......-- ( -19.30, z-score = 0.16, R) >droMoj3.scaffold_6496 19846035 88 + 26866924 -----UACAGAUACACACUUGUCGGCUU-UGCAGGUAAAAAUUUGUAUCUGUUGGACUUUGGAUUUGGUUGCCUGUUGCCAGGUGCCUUGGCGG------ -----.(((((((((.((((((......-.)))))).......)))))))))..(.((..(((((((((........))))))).))..)))..------ ( -22.80, z-score = 0.03, R) >consensus _________AUUAUAUAUAGGUUGGUGUACGUACGUA______CGUACCGUAUGCA____GUAUACCGAUGCCUGUUGCCAGGUGCCAACUUGGCUCU__ .................(((((((((...((((((.............))))))................(((((....))))))))))))))....... (-13.31 = -13.99 + 0.69)



| Location | 18,462,168 – 18,462,268 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 57.35 |
| Shannon entropy | 0.66753 |
| G+C content | 0.44433 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -8.41 |
| Energy contribution | -8.72 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18462168 100 - 21146708 CAAGAGCCAAGUUGGCACCUGGCAACAGGCAUCGGUAUAUACGGUACAUACAGUACGUAUACAUAUGUACGUAUACCAACCAACCUACAAUAUAUAUAAA ..........(((((..((((....))))....(((((((...(((((((..(((....))).))))))))))))))..)))))................ ( -27.70, z-score = -2.76, R) >droSim1.chr2R 17052496 79 - 19596830 --AGAGCCAAGUUGGCACCUGGCAACAGGCAUCGGUAUAC----GGUAUACGGUACA------UACGUACGUACACCAACCUAUAUAUAAA--------- --.(((((.....))).((((....))))..))(((....----(((.((((.(((.------...))))))).))).)))..........--------- ( -23.30, z-score = -1.98, R) >droSec1.super_9 1767054 79 - 3197100 --AGAGCUAAGUUGGCACCUGGUCACAGUCAUCGGUAUAC----UGCAUACGGGACG------UACGUACGUACACCAUCCUAUAUAUUAU--------- --.(((((.....)))..(((....)))...))(((.(((----(((.((((...))------)).))).))).)))..............--------- ( -14.70, z-score = 1.24, R) >droMoj3.scaffold_6496 19846035 88 - 26866924 ------CCGCCAAGGCACCUGGCAACAGGCAACCAAAUCCAAAGUCCAACAGAUACAAAUUUUUACCUGCA-AAGCCGACAAGUGUGUAUCUGUA----- ------..((....)).((((....))))...................(((((((((.((((((..((...-.))..)).)))).))))))))).----- ( -22.00, z-score = -2.04, R) >consensus __AGAGCCAAGUUGGCACCUGGCAACAGGCAUCGGUAUAC____UGCAUACGGUACA______UACGUACGUACACCAACCAAUAUAUAAU_________ .....(((.....))).((((....))))....................................................................... ( -8.41 = -8.72 + 0.31)



| Location | 18,462,204 – 18,462,295 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.43 |
| Shannon entropy | 0.41592 |
| G+C content | 0.46071 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -11.04 |
| Energy contribution | -11.48 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18462204 91 - 21146708 AAAAAGAAAAUCAAUACGAGGCAGACACAAGAGCCAAGUUGGCACCUGGCAACAGG-CAUCGGUAUAUACGGUACAUACA-GUACGUAUACAU ................(((.((..........(((.....)))..(((....))))-).)))((((((...((((.....-)))))))))).. ( -22.80, z-score = -3.69, R) >droSim1.chr2R 17052521 80 - 19596830 -AAAAGAAAAUCAAUACGAGGCAG--ACAAGAGCCAAGUUGGCACCUGGCAACAGG-CAUCGGUAUAC----GGUAUACG-GUACAUAC---- -...............(((.((..--......(((.....)))..(((....))))-).)))((((.(----(.....))-))))....---- ( -19.40, z-score = -2.66, R) >droSec1.super_9 1767079 80 - 3197100 -UAAAGAAAAUCAAUACGAGGCAG--ACUAGAGCUAAGUUGGCACCUGGUCACAGU-CAUCGGUAUAC----UGCAUACG-GGACGUAC---- -.............((((.(((.(--(((((.(((.....)))..))))))...))-).((.((((..----...)))).-)).)))).---- ( -17.20, z-score = -0.66, R) >droYak2.chr2R 15931714 80 - 21139217 -AAAAGAAAAUCAAUACGAGGCAG--ACAAGAGCCAAGUUGGCACCUGGCAACAGG-CAUCGGCAUA-----UACAUACAUACUCCUAC---- -..................(((..--......)))..(((((..((((....))))-..)))))...-----.................---- ( -17.50, z-score = -2.28, R) >droAna3.scaffold_13266 12595327 86 + 19884421 -AAAAUAAAAUCAAUACAAGGCAG--ACAAGAGCCAACCUGGCACCUGGCAACAGGCCUUCCCCGCCCCACUCACUCACAUACCUCUAU---- -..................(((.(--..(((.(((.....))).((((....)))).)))..).)))......................---- ( -17.80, z-score = -2.16, R) >consensus _AAAAGAAAAUCAAUACGAGGCAG__ACAAGAGCCAAGUUGGCACCUGGCAACAGG_CAUCGGUAUAC____UACAUACA_GCACCUAC____ ................(((.............(((.....))).((((....))))...)))............................... (-11.04 = -11.48 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:40 2011