
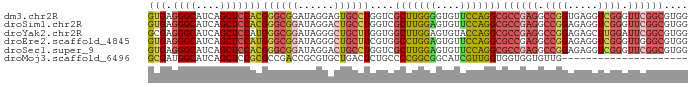
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,444,131 – 18,444,221 |
| Length | 90 |
| Max. P | 0.873071 |

| Location | 18,444,131 – 18,444,221 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Shannon entropy | 0.49821 |
| G+C content | 0.68816 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -30.56 |
| Energy contribution | -31.72 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.37 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18444131 90 + 21146708 GUGAGGGCAUCAGCUCCACGGGCGGAUAGGAGUGCCUGGUCGCUUGGGGUGUUCCAGGCGCCGAGGCCGGUGAGGUCGGGUUCGGCGUGG (((.((((....))))))).........((((..(((.(.....).)))..))))..((((((((.((((.....)))).)))))))).. ( -42.00, z-score = -1.62, R) >droSim1.chr2R 17033918 90 + 19596830 GUGAGGGCAUCAGCUCCACGGGCGGAUAGGACUGCCUGGUCGCUUGGAGUGUUCCAGGCGCCGAGGCCGGAGAGGUCGGGUUCGGCGUGG (((.((((....)))))))((((((......))))))....((((((......))))))((((((.((((.....)))).)))))).... ( -44.10, z-score = -2.04, R) >droYak2.chr2R 15912979 90 + 21139217 GCGAGGGCAUCAGCUCCAUGGGCGGAUAGGGCUGCUUGGUGGCUUGGAGUGUACCAGGCGCCGAGGCCGGAGAGCUUGGAUUCGGCGUGG .((((..((..(((((..(((((((......))))((((((.(((((......)))))))))))..)))..)))))))..))))...... ( -33.30, z-score = 0.80, R) >droEre2.scaffold_4845 15084314 90 - 22589142 GUGAGGGCAUCAGCUCCAUGGGCGGAUAGGGCUGCUUCGUGGCCUGGAGUGUUCCAGGCGCCGAGGCCGGAGAGGUCGGGUUGGGCGUGG ....((((....))))(((((((((......)))).)))))((((((......))))))(((.((.((((.....)))).)).))).... ( -38.80, z-score = -0.60, R) >droSec1.super_9 1749063 90 + 3197100 GUGAGGGCAUCAGCUCCACGGGCGGAUAGGACUGCCUGGUCGCUUGGAGUGUUCCAGGCGCCGAGGCCGGAGAGGUCGGGUUCGGCGUGG (((.((((....)))))))((((((......))))))....((((((......))))))((((((.((((.....)))).)))))).... ( -44.10, z-score = -2.04, R) >droMoj3.scaffold_6496 19813892 69 + 26866924 GCGAUGGCAUCAGCUCCGCGCCGACCGCGUGCUGACUCUGCCCCGGCGGCAUCGUUGGUGGUGGUGUUG--------------------- ((....)).(((((..((((.....)))).)))))(.(..((((((((....)))))).))..).)...--------------------- ( -27.30, z-score = 0.37, R) >consensus GUGAGGGCAUCAGCUCCACGGGCGGAUAGGACUGCCUGGUCGCUUGGAGUGUUCCAGGCGCCGAGGCCGGAGAGGUCGGGUUCGGCGUGG (((.((((....)))))))((((((......))))))....(((((((....)))))))((((((.((((.....)))).)))))).... (-30.56 = -31.72 + 1.16)
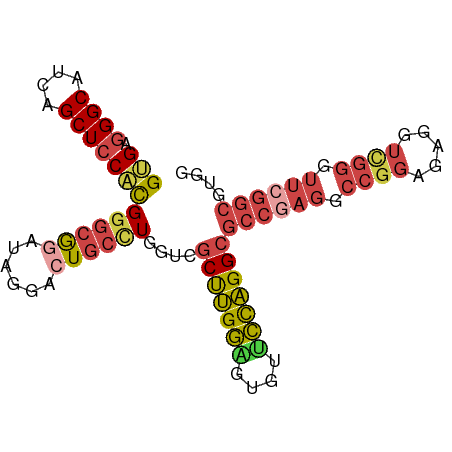

| Location | 18,444,131 – 18,444,221 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Shannon entropy | 0.49821 |
| G+C content | 0.68816 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -15.12 |
| Energy contribution | -16.23 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.583656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18444131 90 - 21146708 CCACGCCGAACCCGACCUCACCGGCCUCGGCGCCUGGAACACCCCAAGCGACCAGGCACUCCUAUCCGCCCGUGGAGCUGAUGCCCUCAC (((((((((..(((.......)))..)))))((((((..............))))))..............)))).((....))...... ( -30.44, z-score = -2.43, R) >droSim1.chr2R 17033918 90 - 19596830 CCACGCCGAACCCGACCUCUCCGGCCUCGGCGCCUGGAACACUCCAAGCGACCAGGCAGUCCUAUCCGCCCGUGGAGCUGAUGCCCUCAC ....(((((..(((.......)))..)))))((.((((....)))).)).....((((.((...((((....))))...))))))..... ( -31.00, z-score = -1.65, R) >droYak2.chr2R 15912979 90 - 21139217 CCACGCCGAAUCCAAGCUCUCCGGCCUCGGCGCCUGGUACACUCCAAGCCACCAAGCAGCCCUAUCCGCCCAUGGAGCUGAUGCCCUCGC ...((((((..((.........))..))))))...(((((((((((.((......)).((.......))...))))).)).))))..... ( -24.10, z-score = -0.04, R) >droEre2.scaffold_4845 15084314 90 + 22589142 CCACGCCCAACCCGACCUCUCCGGCCUCGGCGCCUGGAACACUCCAGGCCACGAAGCAGCCCUAUCCGCCCAUGGAGCUGAUGCCCUCAC ......................(((.((((((((((((....)))))))...............((((....))))))))).)))..... ( -28.10, z-score = -1.63, R) >droSec1.super_9 1749063 90 - 3197100 CCACGCCGAACCCGACCUCUCCGGCCUCGGCGCCUGGAACACUCCAAGCGACCAGGCAGUCCUAUCCGCCCGUGGAGCUGAUGCCCUCAC ....(((((..(((.......)))..)))))((.((((....)))).)).....((((.((...((((....))))...))))))..... ( -31.00, z-score = -1.65, R) >droMoj3.scaffold_6496 19813892 69 - 26866924 ---------------------CAACACCACCACCAACGAUGCCGCCGGGGCAGAGUCAGCACGCGGUCGGCGCGGAGCUGAUGCCAUCGC ---------------------...............(((((.(((....))...((((((.((((.....))))..))))))).))))). ( -23.40, z-score = 0.29, R) >consensus CCACGCCGAACCCGACCUCUCCGGCCUCGGCGCCUGGAACACUCCAAGCGACCAGGCAGCCCUAUCCGCCCGUGGAGCUGAUGCCCUCAC ......................(((.(((((((.(((......))).))...............((((....))))))))).)))..... (-15.12 = -16.23 + 1.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:37 2011