
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,405,155 – 18,405,245 |
| Length | 90 |
| Max. P | 0.579137 |

| Location | 18,405,155 – 18,405,245 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Shannon entropy | 0.33954 |
| G+C content | 0.42957 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.16 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18405155 90 + 21146708 AUAGCAAACUCGAA-AGCAGA--GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGUGG---------- ...(((.((((...-....))--))......(((((...((((......)))).)))))........))).....((((((....))).))).---------- ( -19.80, z-score = -1.35, R) >droSim1.chr2R 16997890 90 + 19596830 AUAGCAAACUCGAA-AGCAGA--GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGUGG---------- ...(((.((((...-....))--))......(((((...((((......)))).)))))........))).....((((((....))).))).---------- ( -19.80, z-score = -1.35, R) >droSec1.super_9 1713414 90 + 3197100 AUAGCAAACUCGAA-AGCAGA--GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGUGG---------- ...(((.((((...-....))--))......(((((...((((......)))).)))))........))).....((((((....))).))).---------- ( -19.80, z-score = -1.35, R) >droYak2.chr2R 15875594 90 + 21139217 AUAGCAAACUCGAA-AGCAGA--GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGGGG---------- ...(((.((((...-....))--))......(((((...((((......)))).)))))........)))(....)..(((....))).....---------- ( -18.90, z-score = -1.38, R) >droEre2.scaffold_4845 15048185 90 - 22589142 AUAGCAAACUCGAA-AGCAGA--GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUUAGUGG---------- .((((.((((((..-.(((..--.((.....(((((...((((......)))).))))).....)).)))(....).))).))).))))....---------- ( -20.90, z-score = -1.89, R) >droAna3.scaffold_13266 12542211 87 - 19884421 ----CUAGCCAGAGGAGCAGA--GAAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGCGA---------- ----...((..((.(((((..--(.......(((((...((((......)))).)))))...........(....).)..))))).)).))..---------- ( -22.00, z-score = -2.18, R) >dp4.chr3 7018612 87 + 19779522 ----------------GAAAGUGGGAGAAUAAGCGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGCGUCGACGGCGAC ----------------....((.(.......(((((...((((......)))).))))).....(((((((....)..(((....))).)))))).).))... ( -22.50, z-score = -1.08, R) >droWil1.scaffold_180745 237470 100 + 2843958 ACAGUAAGAGAGAAAGAGAGACGUGGGCAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGCGUCAACGAC--- ...................(((((.(((...(((((...((((......)))).)))))........((.(....).))......))).)))))......--- ( -22.70, z-score = -1.19, R) >consensus AUAGCAAACUCGAA_AGCAGA__GUAGAAUAAACGCAAAGCUUACACAAAAGUGGCGUUAAAAAUGAUGCGGAAACACGAUGUUCGUCAGUGG__________ ..........................((((((((((...((((......)))).)))))........((.(....).)).))))).................. (-14.15 = -14.16 + 0.02)

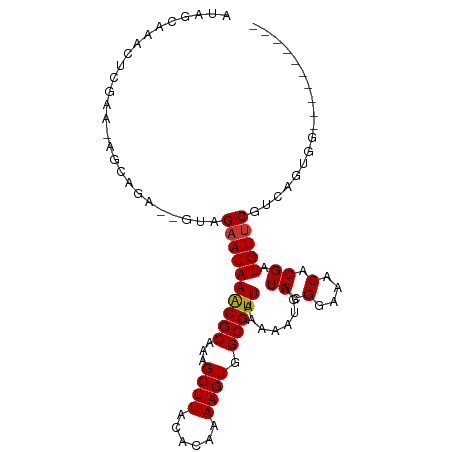
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:32 2011