
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,361,502 – 18,361,612 |
| Length | 110 |
| Max. P | 0.859713 |

| Location | 18,361,502 – 18,361,612 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 54.95 |
| Shannon entropy | 0.94464 |
| G+C content | 0.41610 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -5.15 |
| Energy contribution | -5.12 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18361502 110 - 21146708 -----UUCGUUUCUUUAUUCGAUG-UUUUUCCAAAACGUAUCC-UGUAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACUUCG -----..............(((.(-(...((.((((((....(-(((((.((((....((..(((((((....)))))))..))..)))))))))).))))))...))...)).))) ( -26.90, z-score = -3.65, R) >droEre2.scaffold_4845 15004265 100 + 22589142 --------------UUCUUCGAUG--UUUUCCAAAACGUAUCC-UGUAGGCUUUUCCUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACUUCG --------------.....(((.(--(..((.((((((....(-(((((.((((....((..(((((((....)))))))..))..)))))))))).))))))...))...)).))) ( -26.60, z-score = -3.41, R) >droYak2.chr2R 15830944 114 - 21139217 --UUCUUUUCUUCAUUGUUCGAUGUUUUUUCCAAAACGUAUCC-UGUAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACUUUG --.........((((......(((((((....)))))))...(-(((((.((((....((..(((((((....)))))))..))..))))))))))........))))......... ( -26.80, z-score = -3.43, R) >droSec1.super_9 1669806 113 - 3197100 --UUCGUUUCUUUAUUAUUCGAUG-UUUUUCCAAAACGUAUCC-UGUAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACUUCG --.................(((.(-(...((.((((((....(-(((((.((((....((..(((((((....)))))))..))..)))))))))).))))))...))...)).))) ( -26.90, z-score = -3.57, R) >droSim1.chr2R_random 2778752 106 - 2996586 ------UAUGGUCUUAUAUCGAUG--UUUUUCAAAACGUAUCC-UGUAGACUUUU-UUUGUUUCCCACAU-CUUGUGGGAACCUUCGAAGCUGCAGCUGUUUUUAUGACCUACUUCG ------...((((........(((--(((....))))))...(-(((((.((((.-...(.(((((((..-...))))))).)...))))))))))..........))))....... ( -28.80, z-score = -4.11, R) >droMoj3.scaffold_6496 13125357 92 - 26866924 ------------------UCCGU---CUAUUCA---CCCAUGU-UUUAGCAUGUUGUUUGUGUCCUACAUAAAUGUGGGUGUCCUGGAGGCCAGCGCCGUGGUACUGGCCUACAUUG ------------------....(---(((..((---(((((((-....)).....(((((((.....))))))))))))))...))))((((((..(....)..))))))....... ( -32.30, z-score = -2.55, R) >droVir3.scaffold_12875 816851 114 + 20611582 UCGAUCUUUUGUCAUGUUUUUAU---UUUAACACGGCUCAUACACUUAGCCUGAUUUUUGUGGCCUACGUACAAGUGGGCAUCAUUGAAGCCAGCGCCGUGGUAAUCACCUAUUUUG ..(((.....(((.((((.....---...)))).))).....(((...(((((..(((.(((((((((......))))))..))).)))..))).)).)))...))).......... ( -25.40, z-score = -0.64, R) >droWil1.scaffold_181009 37559 99 - 3585778 ---------------AUUCGAAA---CUGAUCUUAAUGUUUUCUUUUAGUAUAAUAAUGGUGGCUUUUGUAAUUGUGGGAUCCAUUGAGGCAGGAGCUGUCUUUCUUAUAUACUUUA ---------------....((((---(..........)))))......((((((.((.((..((((((((....((((...))))....))))))))..)).)).))))))...... ( -21.60, z-score = -1.46, R) >droAna3.scaffold_13266 12494295 97 + 19884421 ----------------UAUUAUC---UUGUUAAUAAU-UUUAAAUUUAGACUCCUGUUUGUGACACAUAUUUUGGUGGGACCUAUCGAAGCCGCCGCUGUUUUUAUAGUUUACUUUA ----------------(((((..---....)))))..-.........((((....))))((((..........(((((............)))))(((((....))))))))).... ( -11.80, z-score = 0.96, R) >apiMel3.Group5 6900723 87 + 13386189 -------------------------UCCGAUUCUCUA-----UGCACAGGCUUAUUUCGAUGGCGUAUGGUCAGAUCGGUAUGAUCCAGGCGGCCGCCGGUUUCUUCGUCUACUUCG -------------------------.((((((..(((-----(((....(((.........)))))))))...)))))).((((.((.(((....))))).....))))........ ( -19.60, z-score = 1.30, R) >triCas2.ChLG7 16030122 92 - 17478683 -------------------------GCUGACUUUCUAAAUGGUAUGCAGGUUGAUUUCGAUGGCAUACGGCCAGAUUGGUAUGAUUCAAGCAGCUGCUGGUUUCUUCGUGUACUUUG -------------------------((((.(((........((((((..((((....)))).)))))).(((.....))).......)))))))((((((.....))).)))..... ( -20.40, z-score = 0.68, R) >consensus ______________U_UUUCGAU___UUUUCCAAAACGUAUCC_UGUAGACUUUUCUUUGUGUCCCACAUCCUUGUGGGAACCAUCGAAGCUGCAGCUGUUUUUAUGACCUACUUUG ..........................................................................(((((.........(((....)))..........))))).... ( -5.15 = -5.12 + -0.03)

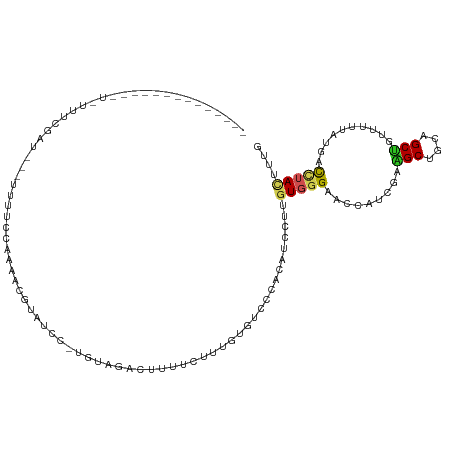
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:25 2011