
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,357,597 – 18,357,693 |
| Length | 96 |
| Max. P | 0.521246 |

| Location | 18,357,597 – 18,357,693 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 61.34 |
| Shannon entropy | 0.77460 |
| G+C content | 0.65920 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.40 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18357597 96 + 21146708 ------GCCGCAGCGGCUGCCUGCGGGGAGCAUCUCUCUGGAC-AGACGACUCCGGCGACGGCGGCGGCA---UGUAAGUGUACUCC-----GGCCGCAGUGGCUUUGCCA ------(((((.(((((((.((((((((((...)))))))..)-)).((.(.(((....))).).))(((---(....))))....)-----)))))).)))))....... ( -40.70, z-score = 0.58, R) >droSec1.super_9 1665955 96 + 3197100 ------GCCGCAGCGGCUGCUUGCGGGGAGCAUCUCUCCGGAC-AGGCGACUCCGGCGACGGCGGCGGCA---CGUAAGUCUACUCC-----GGCCGUAGUGGCUUUGCCA ------(((((.(((((((....(((((((...)))))))(((-..(((...(((.((....)).)))..---)))..))).....)-----)))))).)))))....... ( -44.80, z-score = -0.52, R) >droYak2.chr2R 15827007 79 + 21139217 ------GCCGCAGCGGCUGCCUGCGGGGAGCAUCUCUCCGGAC-AGGCGACUCCGGCGACGGCGGCGGCA---CGUAAGUGUACUCC-----GG----------------- ------((((..((((.(((((((((((((...)))))))..)-)))))...)).))..))))((..(((---(....))))...))-----..----------------- ( -39.50, z-score = -1.27, R) >droEre2.scaffold_4845 15000311 96 - 22589142 ------GCCGCAGCUGCUGCCUGCGGGGAGCAUCUCUCCGGGC-AGGCGACUCCGGCGACGGCAGCGGCA---UGUAAGUGUACUCC-----GGCCAUAGUGGCUAUGCCA ------(((((.((((((((((((((((((...))))))..))-)))))....((....)))))))(((.---.(((....)))...-----.)))...)))))....... ( -45.60, z-score = -0.61, R) >droGri2.scaffold_15245 6907188 108 + 18325388 GCUGCUGCAUCCGCCAUGGCCUGUGGGGACCAUCUCGCUGGGC-AGGCGAAUUCGGCGACGGCAGCAGCGGCUCGUGAGUAUAUCCUAGUUAAAAAGGAAAAAUUACCG-- ((((((((...((((.((.((.(((((......))))).)).)-)))))....((....))))))))))((..(....)....((((........)))).......)).-- ( -43.90, z-score = -2.12, R) >droVir3.scaffold_12875 812635 104 - 20611582 GCUGCAGCUGCGGCUGCCGCCUGUGGGGAGCAUCUCACUGGAC-AGGCGAAUACUGCGACAGCAGCAGCA---CGUGAGUAU-CUCCAGCAGGGCAAAUGUAAUUACCG-- ..(((..((((.((((((((((((.(((((...))).)).).)-))))).....(((....))))))))(---(....))..-.....)))).))).............-- ( -38.10, z-score = -0.00, R) >droAna3.scaffold_13266 12490479 99 - 19884421 ---GCAGCGGCAGCGGCUGCCUGCGGAGAGCAUCUCACGGGAC-AGGCGACACCGGCGACGGCGGCAGCA---CGUAAGUGGACUUCUCUUCGGCAGCAGUGGUUU----- ---((....))(((.(((((.(((((((((..((.(((...((-..((..(.(((....))).)...)).---.))..)))))...)))))).)))))))).))).----- ( -38.50, z-score = 0.07, R) >droSim1.chr2R 16904738 100 + 19596830 ------GUCAGUGCGGCCAGCUUCAAUGGCCGAAACGCCGGGCUGGGUAAUGCUGGCGGUGGCGGAAUCA--UGGCCAAUAGCAUCGUGGCCACCACUGGCGGUGGCA--- ------((((...((((((.......))))))...((((((((..(......)..))((((((.(.....--..((.....))....).)))))).)))))).)))).--- ( -45.10, z-score = -0.80, R) >consensus ______GCCGCAGCGGCUGCCUGCGGGGAGCAUCUCUCCGGAC_AGGCGACUCCGGCGACGGCGGCAGCA___CGUAAGUGUACUCC_____GGCAGUAGUGGUUACG___ ......(((((.((....((((.(((((((...)))))))....)))).....((....)))).))))).......................................... (-17.10 = -18.40 + 1.30)
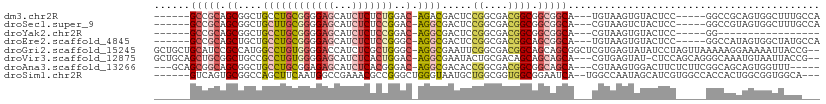
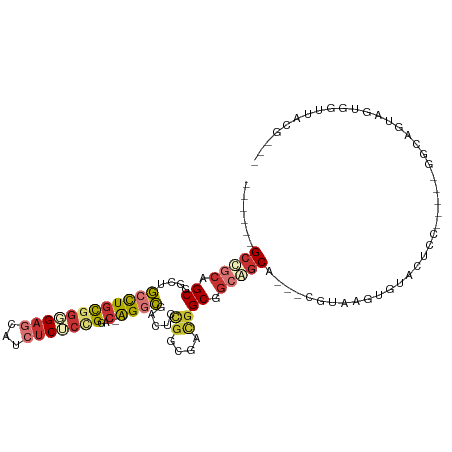

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:24 2011