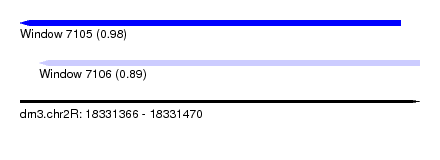
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,331,366 – 18,331,470 |
| Length | 104 |
| Max. P | 0.982040 |

| Location | 18,331,366 – 18,331,465 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Shannon entropy | 0.14587 |
| G+C content | 0.62041 |
| Mean single sequence MFE | -46.07 |
| Consensus MFE | -42.01 |
| Energy contribution | -42.90 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.982040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
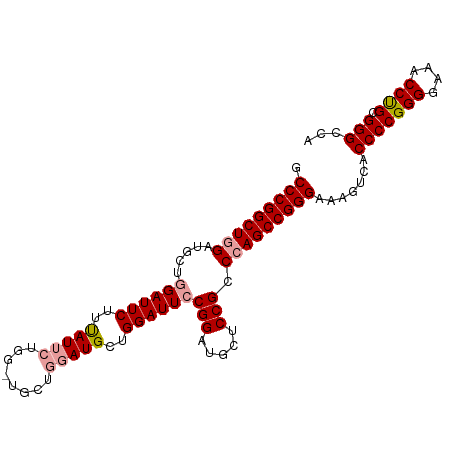
>dm3.chr2R 18331366 99 - 21146708 GCCCGGCUGGAUGCUGGAUUCUUUAUUCUGGAUGCUGGAUGUUGGAUUGCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCCGCGGGCCA .(((((((((......(((((..(((((........)))))..)))))((((.....))))))))))))).......(((((((....)))).)))... ( -50.50, z-score = -3.15, R) >droSim1.chr2R 16931538 99 - 19596830 GCCCGGCUGGAUUCUGGAUUCUUUAUUCUGGCUGCUGGAUGCUGGAUUCCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCUGCGGGCCA .(((((((((..((((((.(((....((..(...)..))....))).)))))).((...))))))))))).......(((((((....)))).)))... ( -47.20, z-score = -2.31, R) >droSec1.super_9 1639916 92 - 3197100 GCCCGGCUGGAUGCUGGAUUCUUCAUU-------CUGUAUGCUGGAUUCCGGAUGCUCCGCCAAGCCGGGAAAGUCACCCCGGGGAAACCUGUGGGCCA .((((((((((..(((((.(((.(((.-------....)))..))).)))))....)))....))))))).......(((((((....)))).)))... ( -40.50, z-score = -1.82, R) >consensus GCCCGGCUGGAUGCUGGAUUCUUUAUUCUGG_UGCUGGAUGCUGGAUUCCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCUGCGGGCCA .(((((((((.....((((((..(((((........)))))..))))))(((.....))).))))))))).......(((((((....)))).)))... (-42.01 = -42.90 + 0.89)

| Location | 18,331,371 – 18,331,470 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Shannon entropy | 0.14587 |
| G+C content | 0.61005 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -35.87 |
| Energy contribution | -36.77 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
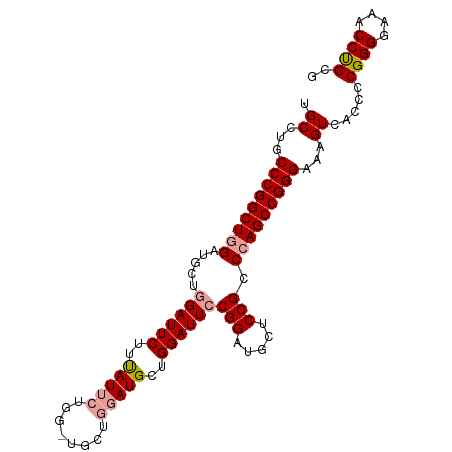
>dm3.chr2R 18331371 99 - 21146708 UGCCUGCCCGGCUGGAUGCUGGAUUCUUUAUUCUGGAUGCUGGAUGUUGGAUUGCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCCGCG .((...(((((((((......(((((..(((((........)))))..)))))((((.....)))))))))))))...........(((....))))). ( -44.70, z-score = -2.59, R) >droSim1.chr2R 16931543 99 - 19596830 UGCCUGCCCGGCUGGAUUCUGGAUUCUUUAUUCUGGCUGCUGGAUGCUGGAUUCCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCUGCG .((...(((((((((..((((((.(((....((..(...)..))....))).)))))).((...)))))))))))...)).....((((....)))).. ( -41.00, z-score = -1.63, R) >droSec1.super_9 1639921 92 - 3197100 UGCCUGCCCGGCUGGAUGCUGGAUUCUUCAUU-------CUGUAUGCUGGAUUCCGGAUGCUCCGCCAAGCCGGGAAAGUCACCCCGGGGAAACCUGUG .((...((((((((((..(((((.(((.(((.-------....)))..))).)))))....)))....)))))))...)).....((((....)))).. ( -34.50, z-score = -1.15, R) >consensus UGCCUGCCCGGCUGGAUGCUGGAUUCUUUAUUCUGG_UGCUGGAUGCUGGAUUCCGGAUGCUCCGCCCAGCCGGGAAAGUCACCCCGGGGAAACCUGCG .((...(((((((((.....((((((..(((((........)))))..))))))(((.....))).)))))))))...)).....((((....)))).. (-35.87 = -36.77 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:20 2011