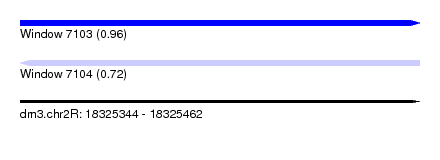
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,325,344 – 18,325,462 |
| Length | 118 |
| Max. P | 0.955228 |

| Location | 18,325,344 – 18,325,462 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.47481 |
| G+C content | 0.47018 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -18.50 |
| Energy contribution | -17.98 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.955228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
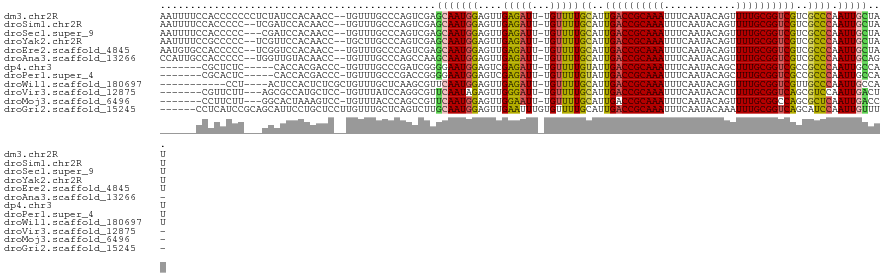
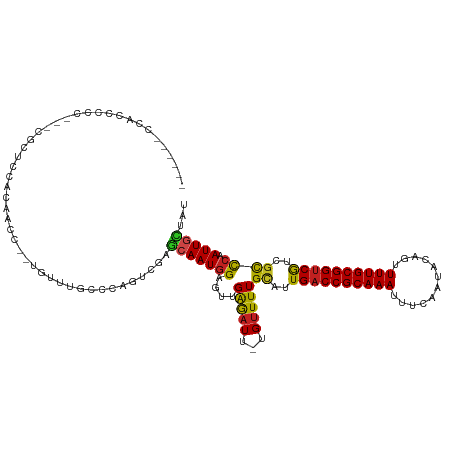
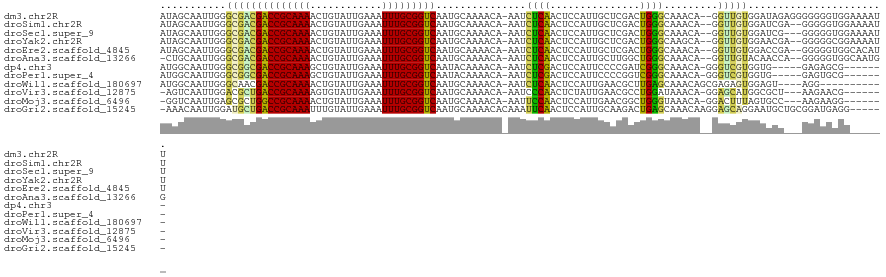
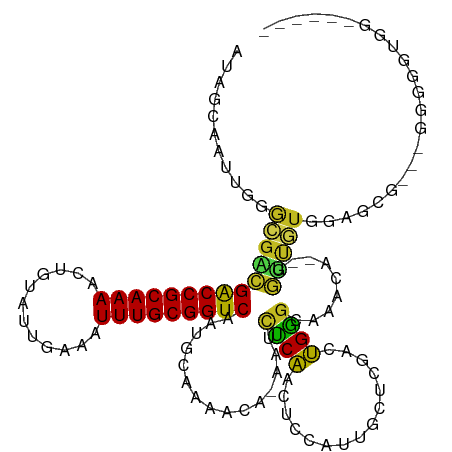
>dm3.chr2R 18325344 118 + 21146708 AAUUUUCCACCCCCCCUCUAUCCACAACC--UGUUUGCCCAGUCGAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU ...............(((..((((.....--((((((......)))))).))))...)))...-......((..((((((((((............))))))))))..))........... ( -26.40, z-score = -2.03, R) >droSim1.chr2R 16925571 116 + 19596830 AAUUUUCCACCCCC--UCGAUCCACAACC--UGUUUGCCCAGUCGAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU .............(--((((((((.....--((((((......)))))).)))).)))))...-......((..((((((((((............))))))))))..))........... ( -27.60, z-score = -1.89, R) >droSec1.super_9 1633998 115 + 3197100 AAUUUUCCACCCCC---CGAUCCACAACC--UGUUUGCCCAGUCGAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU ..............---((((...(((..--...)))....))))((((((((....((((..-..))))((..((((((((((............))))))))))..)))).)))))).. ( -25.30, z-score = -1.28, R) >droYak2.chr2R 15794733 116 + 21139217 AAUUUUCCGCCCCC--UCGUUCCACAACC--UGCUUGCCCAGUCGAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU ........((...(--((..((((.....--((((((......)))))).))))...)))...-......((..((((((((((............))))))))))..))......))... ( -28.10, z-score = -1.66, R) >droEre2.scaffold_4845 14968926 116 - 22589142 AAUGUGCCACCCCC--UCGGUCCACAACC--UGUUUGCCCAGUCGAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU ..((((..(((...--..))).))))..(--((......)))...((((((((....((((..-..))))((..((((((((((............))))))))))..)))).)))))).. ( -28.70, z-score = -0.82, R) >droAna3.scaffold_13266 12454962 115 - 19884421 CCAUUGCCACCCCC--UGGUUGUACAACC--UGUUUGCCCAGCCAAGCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCAG- (((((((.......--((((((..(((..--...)))..)))))).)))))))((((((((..-...)))((..((((((((((............))))))))))..)).)))))....- ( -32.90, z-score = -2.24, R) >dp4.chr3 8925938 107 + 19779522 -------CGCUCUC-----CACCACGACCC-UGUUUGCCCGAUCGGGGAAUGGAGUCGAGAUU-UGUUUUGUAUUGACCGCAAAUUUCAAUACAGCUUUGCGGUCGCCGCCCAAUUGCCAU -------.((....-----.....((((.(-(((((.(((....))))))))).)))).....-......((..((((((((((............))))))))))..))......))... ( -28.90, z-score = -0.46, R) >droPer1.super_4 4247087 107 + 7162766 -------CGCACUC-----CACCACGACCC-UGUUUGCCCGACCGGGGAAUGGAGUCGAGAUU-UGUUUUGUAUUGACCGCAAAUUUCAAUACAGCUUUGCGGUCGCCGCCCAAUUGCCAU -------.(((...-----.....((((.(-(((((.(((....))))))))).)))).....-......((..((((((((((............))))))))))..)).....)))... ( -30.00, z-score = -0.89, R) >droWil1.scaffold_180697 4145710 105 - 4168966 -----------CCU----ACUCCACUCUCGCUGUUUGCUCAAGCGUUCAAUGGAGUUGAGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUUGCCCAAUUGCCAU -----------...----.(((.((((.((.((..(((....)))..)).)))))).)))...-......(((.((((((((((............)))))))))).)))........... ( -31.90, z-score = -3.21, R) >droVir3.scaffold_12875 19370372 108 + 20611582 -------CGUUCUU---AGCGCCAUGCUCC-UGUUUAUCCAGGCGUUCAAUAGAGUUGGGAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACACUUUUGCGGUCAGCGUCCAAUUGACU- -------...((((---(((.....((.((-((......)))).))........)))))))..-......((..((((((((((............))))))))))))(((.....))).- ( -29.02, z-score = -1.58, R) >droMoj3.scaffold_6496 13072017 108 + 26866924 -------CCUUCUU---GGCACUAAAGUCC-UGUUUACCCAGCCGUUCAAUGGAGUUGGAAUU-UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGCCAGCGCUCAAUUGACC- -------.......---((((....(((.(-((........(((((..(((.(.(((((((((-(((............)))))))))))).).)))..)))))))).)))....)).))- ( -23.10, z-score = 0.28, R) >droGri2.scaffold_15245 6869233 114 + 18325388 ------CCUCAUCCGCAGCAUUCCUGCUCCUUGUUUGCUCAGUCUUGCAAUGGAGUUGAAUUUGUGUUUUGCAUUGACCGCAAAUUUCAAUACAAAUUUGCGGUCAGCAUCCAAUUGUUU- ------.......(((((.((((..(((((....((((........)))).))))).)))))))))...(((..(((((((((((((......))))))))))))))))...........- ( -37.60, z-score = -4.47, R) >consensus ______CCACCCCC___CGCUCCACAACC__UGUUUGCCCAGUCGAGCAAUGGAGUUGAGAUU_UGUUUUGCAUUGACCGCAAAUUUCAAUACAGUUUUGCGGUCGUCGCCCAAUUGCUAU ..............................................(((((((....(((((...)))))((..((((((((((............))))))))))..)))).)))))... (-18.50 = -17.98 + -0.52)
| Location | 18,325,344 – 18,325,462 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Shannon entropy | 0.47481 |
| G+C content | 0.47018 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -17.99 |
| Energy contribution | -16.88 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.716626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18325344 118 - 21146708 AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUCGACUGGGCAAACA--GGUUGUGGAUAGAGGGGGGGUGGAAAAUU ...(((....(....)(((((((((............)))))))))..)))......-.(((((..(((.(((.(.(((((.(.....).--))))).)))).)))..)))))........ ( -31.70, z-score = -1.26, R) >droSim1.chr2R 16925571 116 - 19596830 AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUCGACUGGGCAAACA--GGUUGUGGAUCGA--GGGGGUGGAAAAUU ...(((....(....)(((((((((............)))))))))..)))......-....((.(((((.((((((....))))))...--((((...))))..--.))))).))..... ( -30.50, z-score = -0.92, R) >droSec1.super_9 1633998 115 - 3197100 AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUCGACUGGGCAAACA--GGUUGUGGAUCG---GGGGGUGGAAAAUU ...(((....(....)(((((((((............)))))))))..)))......-.........((((((.(((((((..((((...--..)))))).)))---)).))))))..... ( -33.00, z-score = -1.69, R) >droYak2.chr2R 15794733 116 - 21139217 AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUCGACUGGGCAAGCA--GGUUGUGGAACGA--GGGGGCGGAAAAUU ...((..(((.(((..(((((((((............)))))))))..)))......-..((((((((...((((((....))))))...--))))).))).)))--....))........ ( -32.30, z-score = -1.19, R) >droEre2.scaffold_4845 14968926 116 + 22589142 AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUCGACUGGGCAAACA--GGUUGUGGACCGA--GGGGGUGGCACAUU ...((..(((.(((..(((((((((............)))))))))..)))....))-)......(((((.((((((....))))))...--((((...))))..--.))))).))..... ( -33.80, z-score = -1.16, R) >droAna3.scaffold_13266 12454962 115 + 19884421 -CUGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGCUUGGCUGGGCAAACA--GGUUGUACAACCA--GGGGGUGGCAAUGG -.((((....(....)(((((((((............)))))))))..)))).....-.....((...(((((.(((((.((..(((...--..)))..)).)))--)).)))))...)). ( -35.00, z-score = -1.43, R) >dp4.chr3 8925938 107 - 19779522 AUGGCAAUUGGGCGGCGACCGCAAAGCUGUAUUGAAAUUUGCGGUCAAUACAAAACA-AAUCUCGACUCCAUUCCCCGAUCGGGCAAACA-GGGUCGUGGUG-----GAGAGCG------- ...((......)).(((((((((((.(......)...)))))))))...........-........(((((..((.(((((.(.....).-.))))).))))-----))).)).------- ( -33.20, z-score = -0.75, R) >droPer1.super_4 4247087 107 - 7162766 AUGGCAAUUGGGCGGCGACCGCAAAGCUGUAUUGAAAUUUGCGGUCAAUACAAAACA-AAUCUCGACUCCAUUCCCCGGUCGGGCAAACA-GGGUCGUGGUG-----GAGUGCG------- ...((......)).(((((((((((.(......)...)))))))))...........-.......((((((..((.((..(.(.....).-.)..)).))))-----)))))).------- ( -33.00, z-score = -0.27, R) >droWil1.scaffold_180697 4145710 105 + 4168966 AUGGCAAUUGGGCAACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCUCAACUCCAUUGAACGCUUGAGCAAACAGCGAGAGUGGAGU----AGG----------- ...(((....(....)(((((((((............)))))))))..)))......-.......((((((((...((((.(......)))))..))))))))----...----------- ( -30.90, z-score = -2.31, R) >droVir3.scaffold_12875 19370372 108 - 20611582 -AGUCAAUUGGACGCUGACCGCAAAAGUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUCCCAACUCUAUUGAACGCCUGGAUAAACA-GGAGCAUGGCGCU---AAGAACG------- -.((...((((.(((((((((((((..(......)..))))))))).((((......-.........((....))...((((......))-)).))))))))))---))..)).------- ( -31.30, z-score = -2.58, R) >droMoj3.scaffold_6496 13072017 108 - 26866924 -GGUCAAUUGAGCGCUGGCCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA-AAUUCCAACUCCAUUGAACGGCUGGGUAAACA-GGACUUUAGUGCC---AAGAAGG------- -((........(((.((((((((((............)))))))))).)))......-...........(((((((.(.(((......))-)..))))))))))---.......------- ( -25.60, z-score = 0.16, R) >droGri2.scaffold_15245 6869233 114 - 18325388 -AAACAAUUGGAUGCUGACCGCAAAUUUGUAUUGAAAUUUGCGGUCAAUGCAAAACACAAAUUCAACUCCAUUGCAAGACUGAGCAAACAAGGAGCAGGAAUGCUGCGGAUGAGG------ -...........((((((((((((((((......)))))))))))))..))).........((((..(((.((((........))))....(.((((....)))).)))))))).------ ( -34.60, z-score = -3.10, R) >consensus AUAGCAAUUGGGCGACGACCGCAAAACUGUAUUGAAAUUUGCGGUCAAUGCAAAACA_AAUCUCAACUCCAUUGCUCGACUGGGCAAACA__GGUUGUGGAGCG___GGGGGUGG______ ...........((((((((((((((............)))))))))...............((((...............)))).........)))))....................... (-17.99 = -16.88 + -1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:19 2011