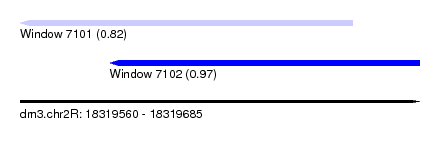
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,319,560 – 18,319,685 |
| Length | 125 |
| Max. P | 0.965341 |

| Location | 18,319,560 – 18,319,664 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.40 |
| Shannon entropy | 0.61988 |
| G+C content | 0.48252 |
| Mean single sequence MFE | -29.61 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.08 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
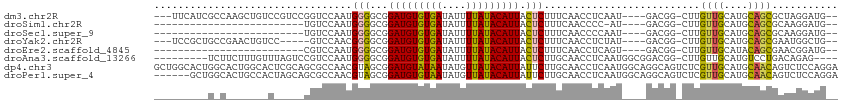
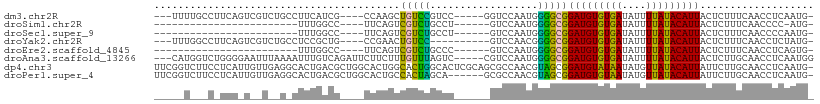
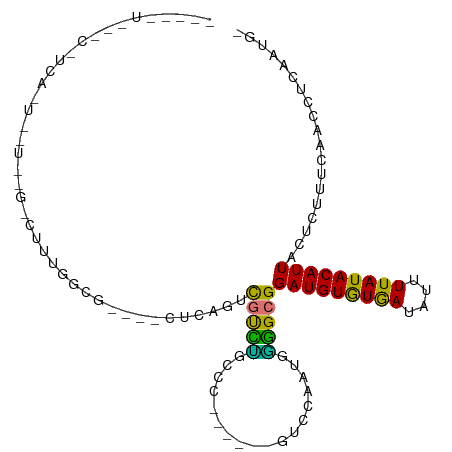
>dm3.chr2R 18319560 104 - 21146708 ---UUCAUCGCCAAGCUGUCCGUCCGGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAAU----GACGG-CUUGUUGCAUGCAGCGCUAGGAUG-- ---..((((...(((((((((((((.((...)).)))))(((((((((....))))))))).................----)))))-)))((((....)))).....))))-- ( -32.70, z-score = -1.56, R) >droSim1.chr2R 16919867 81 - 19596830 -------------------------UGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCCC-AU----GACGG-CUUGUUGCAUGCAGCGCAAGGAUG-- -------------------------.((((.((((((..(((((((((....)))))))........))..))))-))----....(-(..((((....))))))..)))).-- ( -24.60, z-score = -1.35, R) >droSec1.super_9 1628040 82 - 3197100 -------------------------UGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCCCAAU----GACGG-CUUGUUGCAUGCAGCGCAAGGAUG-- -------------------------.((((..(((((..(((((((((....)))))))........))..)))))..----....(-(..((((....))))))..)))).-- ( -23.60, z-score = -1.04, R) >droYak2.chr2R 15788502 99 - 21139217 ---UCCGCUGCCGAACUGUCC-----GUCCAACGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCUAU----GACGG-CUUGUUGCAUGCAGCGAAUGGCUG-- ---...(((((((....((((-----((((....))))))))((((((....))))))....................----..)))-)((((((....))))))..)))..-- ( -32.30, z-score = -2.36, R) >droEre2.scaffold_4845 14962950 82 + 22589142 -------------------------CGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAGU----GACGG-CUUGUUGCAUACAGCGAACGGAUG-- -------------------------(((((..(((((..(((((((((....)))))))........))..)))))..----....(-.((((((....)))))).))))))-- ( -21.40, z-score = -0.99, R) >droAna3.scaffold_13266 12448890 100 + 19884421 ---------UCUUCUUUGUUUAGUCCGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUGCAACCUCAAUGGCGGACGG-CUUGUUGCAUGUCCUGACAGAG---- ---------....(((((((..(((((.(((.(((((..(((((((((....)))))))))..........))))).)))))))).(-(.....)).......)))))))---- ( -32.50, z-score = -2.59, R) >dp4.chr3 8920184 114 - 19779522 GCUGGCACUGGCACUGGCACUCGCAGCGCCAACGUAGCGGAUGUAUAAUAUGUUAUACAUUAUUCUUGCAACCUCAAUGGCAGGCAGUCUCGUUGCAUGCAACAGUCUCCAGGA .((((.(((((((.((((.(.....).))))..(((((((((((((((....)))))))))....((((..((.....))...))))...)))))).)))..))))..)))).. ( -36.00, z-score = -0.69, R) >droPer1.super_4 4241344 108 - 7162766 ------GCUGGCACUGCCACUAGCAGCGCCAACGUAGCGGAUGUGUAAUAUGUUAUACAUUAUUCUUGCAACCUCAAUGGCAGGCAGUCUCGUUGCAUGCAACAGUCUCCAGGA ------((((((.((((.....)))).))))..(((((((((((((((....)))))))))....((((..((.....))...))))...))))))..)).............. ( -33.80, z-score = -0.69, R) >consensus _________G_C_______C_____GGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAAU____GACGG_CUUGUUGCAUGCAGCGAAAGGAUG__ .................................(((...(((((((((....))))))))).)))..........................((((....))))........... (-12.62 = -12.08 + -0.54)
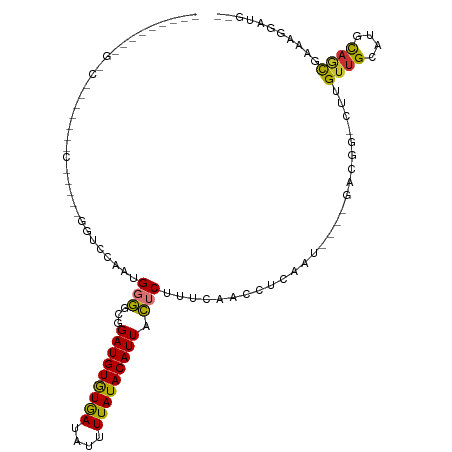
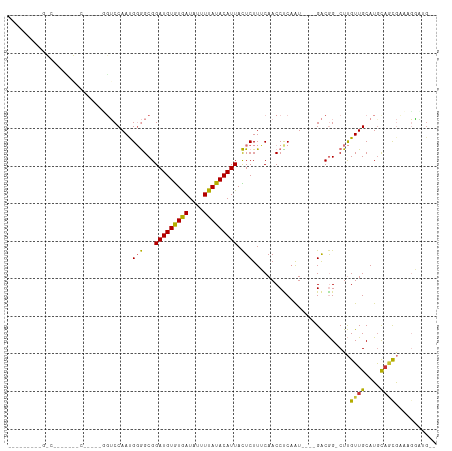
| Location | 18,319,588 – 18,319,685 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 61.24 |
| Shannon entropy | 0.73480 |
| G+C content | 0.46348 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -9.94 |
| Energy contribution | -8.53 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18319588 97 - 21146708 ---UUUUGCCUUCAGUCGUCUGCCUUCAUCG----CCAAGCUGUCCGUCC-----GGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAAUG- ---........(((..(((((((((.(((.(----((..((.....))..-----)))...))))))))))))..)))...............................- ( -19.90, z-score = -0.30, R) >droSim1.chr2R 16919895 74 - 19596830 ------------------------UUUGGCC----UUCAGUCGUCUGCCU------GUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCCC-AUG- ------------------------..(((.(----..(((....)))...------).)))((((((..(((((((((....)))))))........))..))))-)).- ( -19.50, z-score = -1.61, R) >droSec1.super_9 1628068 75 - 3197100 ------------------------UUUGGCC----UUCAGUCGUCUGCCU------GUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCCCAAUG- ------------------------.((((.(----..(((....)))...------).))))(((((..(((((((((....)))))))........))..)))))...- ( -19.20, z-score = -1.45, R) >droYak2.chr2R 15788530 92 - 21139217 ---UUUGGCCUUCAGUCGUCUGCCUCCGCUG----CCGAACUGUCC----------GUCCAACGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCUAUG- ---((((((....((.((........)))))----)))))..((((----------((((....))))))))((((((....)))))).....................- ( -22.80, z-score = -1.29, R) >droEre2.scaffold_4845 14962978 75 + 22589142 ------------------------UUUGGCC----UUCAGUCGUCUGCCC------GUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAGUG- ------------------------...(((.----....)))((((((((------........))))))))((((((....)))))).....................- ( -19.00, z-score = -1.30, R) >droAna3.scaffold_13266 12448919 102 + 19884421 ---CAUGGUCUGGGGAAUUUAAAAUUUGUCAGAUUCUUCUUUGUUUAGUC-----CGUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUGCAACCUCAAUGG ---((((((((((.((((.....)))).))))))).....((((...(((-----(((((....))))))))((((((....)))))).........)))).....))). ( -26.70, z-score = -1.74, R) >dp4.chr3 8920219 109 - 19779522 UUCGGUCUUCCUCAUUGUUGAGGCACUGACGCUGGCACUGGCACUGGCACUCGCAGCGCCAACGUAGCGGAUGUAUAAUAUGUUAUACAUUAUUCUUGCAACCUCAAUG- .(((((...(((((....))))).)))))(((((.(..((((.(((.......))).))))..))))))(((((((((....)))))))))..................- ( -33.30, z-score = -1.85, R) >droPer1.super_4 4241379 103 - 7162766 UUCGGUCUUCCUCAUUGUUGAGGCACUGACGCUGGCACUGCCACUAGCA------GCGCCAACGUAGCGGAUGUGUAAUAUGUUAUACAUUAUUCUUGCAACCUCAAUG- ...((....)).....(((((((.....(((.((((.((((.....)))------).)))).))).((((((((((((....))))))))).....)))..))))))).- ( -33.90, z-score = -2.79, R) >consensus _____U___C_UCA_U__U__G_CUUUGGCG____CUCAGUCGUCUGCCC______GUCCAAUGGGGCGGAUGUGUGAUAUUUUAUACAUUACUCUUUCAACCUCAAUG_ .........................................(((((..................)))))(((((((((....)))))))))................... ( -9.94 = -8.53 + -1.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:17 2011