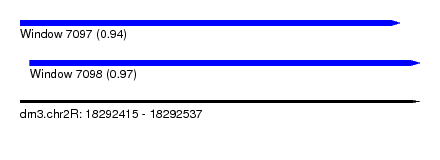
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,292,415 – 18,292,537 |
| Length | 122 |
| Max. P | 0.971228 |

| Location | 18,292,415 – 18,292,531 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.21 |
| Shannon entropy | 0.48630 |
| G+C content | 0.34842 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -14.39 |
| Energy contribution | -15.71 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.938099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
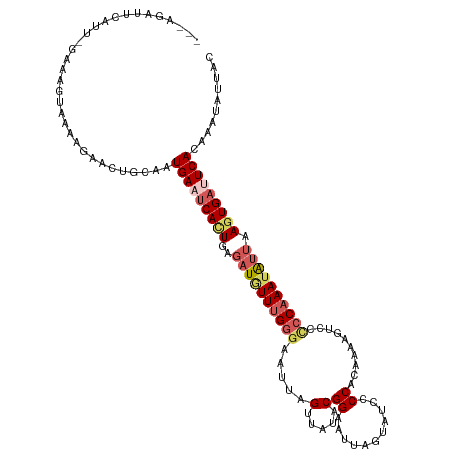
>dm3.chr2R 18292415 116 + 21146708 ---GGAUUCAUU-GGCAGUAGAAGAACUGCAAUGAUUCACUGAUAUGUUUGGGAAUCAGUUACACGGAAUUAGUAUAUCCACAAAAGUUCCCCAAAUAUUAAUUGAUUCACAAACAUUAC ---.((.(((((-(.((((......)))))))))).))......(((((((.((((((((((...((((((.((......))...))))))........)))))))))).)))))))... ( -30.90, z-score = -3.50, R) >droSim1.chr2R 16892194 116 + 19596830 ---GGAUUCAUU-GGCAGUAAAAGAACUGCAAUGAAUCACUGAGAUCUUUGGGAAUUAGUUAUACGGAAUAAGUAUCCCCACAAAAGUCCCCCAAAUGUUAAGUGAUUCAAAGAUAUUAC ---.........-.(((((......)))))..(((((((((..(((.((((((............(((.......)))............)))))).))).))))))))).......... ( -28.15, z-score = -2.41, R) >droSec1.super_9 1600437 100 + 3197100 --------------------AAGGAACUGCAAUGAAUCACUGAGAUGUUUGGGUAUUAGUUAUACGGAAUAAGUAUCCCCACAAAAGUCCCCCAAAUGUUAAGUGAUUCAAAGAUAUUAC --------------------............(((((((((..((((((((((............(((.......)))............)))))))))).))))))))).......... ( -24.35, z-score = -2.55, R) >droYak2.chr2R 18042089 111 + 21139217 ---AGAUUCAUUCGAAAGUAAAAGAACUGGCAUGAAUCAUUGGGAUGUUUGGGAAUUAGUUAUACGGAAUUAGUAUCUCC---AGGUUCUUCCAAAUAUUAAGUGAUUCACAAAUAU--- ---..........(..(((......)))..).(((((((((..((((((((((((((.(..((((.......))))..).---.)))))..))))))))).))))))))).......--- ( -24.70, z-score = -1.50, R) >droEre2.scaffold_4845 12432864 114 + 22589142 AGAACUUCCAUUUGAAAGUAAGGGAACUGUUGUGAAGCAUUAAGUUAUUCGUAAACAAGUUAUCCGGAAUUAGUAUCCCC---AGGUUACCCCAAAUAUUAAGUGAAUCACAAAGAU--- ........(((((((......(((((((.(((((((((.....))..)))))))...))))....(((.......)))..---......)))......)))))))............--- ( -13.90, z-score = 1.47, R) >consensus ___AGAUUCAUU_GAAAGUAAAAGAACUGCAAUGAAUCACUGAGAUGUUUGGGAAUUAGUUAUACGGAAUUAGUAUCCCCACAAAAGUCCCCCAAAUAUUAAGUGAUUCACAAAUAUUAC ................................(((((((((..((((((((((............(((.......)))............)))))))))).))))))))).......... (-14.39 = -15.71 + 1.32)

| Location | 18,292,418 – 18,292,537 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.14 |
| Shannon entropy | 0.50334 |
| G+C content | 0.34470 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -14.39 |
| Energy contribution | -15.71 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
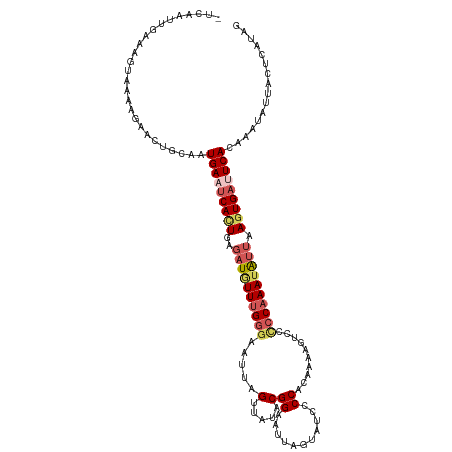
>dm3.chr2R 18292418 119 + 21146708 -UUCAUUGGCAGUAGAAGAACUGCAAUGAUUCACUGAUAUGUUUGGGAAUCAGUUACACGGAAUUAGUAUAUCCACAAAAGUUCCCCAAAUAUUAAUUGAUUCACAAACAUUACUCAUAG -.((((((.((((......)))))))))).....(((.(((((((.((((((((((...((((((.((......))...))))))........)))))))))).)))))))...)))... ( -29.80, z-score = -3.44, R) >droSim1.chr2R 16892197 119 + 19596830 -UUCAUUGGCAGUAAAAGAACUGCAAUGAAUCACUGAGAUCUUUGGGAAUUAGUUAUACGGAAUAAGUAUCCCCACAAAAGUCCCCCAAAUGUUAAGUGAUUCAAAGAUAUUACUCAUAG -.......(((((......)))))..(((((((((..(((.((((((............(((.......)))............)))))).))).)))))))))................ ( -28.15, z-score = -2.60, R) >droSec1.super_9 1600437 106 + 3197100 --------------AAGGAACUGCAAUGAAUCACUGAGAUGUUUGGGUAUUAGUUAUACGGAAUAAGUAUCCCCACAAAAGUCCCCCAAAUGUUAAGUGAUUCAAAGAUAUUACUCAUAG --------------............(((((((((..((((((((((............(((.......)))............)))))))))).)))))))))................ ( -24.35, z-score = -2.04, R) >droYak2.chr2R 18042092 108 + 21139217 UUCAUUCGAAAGUAAAAGAACUGGCAUGAAUCAUUGGGAUGUUUGGGAAUUAGUUAUACGGAAUUAGUAUCUCCA---GGUUCUUCCAAAUAUUAAGUGAUUCACAAAUAU--------- .......(..(((......)))..).(((((((((..((((((((((((((.(..((((.......))))..)..---)))))..))))))))).))))))))).......--------- ( -24.70, z-score = -1.86, R) >droEre2.scaffold_4845 12432870 108 + 22589142 UCCAUUUGAAAGUAAGGGAACUGUUGUGAAGCAUUAAGUUAUUCGUAAACAAGUUAUCCGGAAUUAGUAUCCCCA---GGUUACCCCAAAUAUUAAGUGAAUCACAAAGAU--------- ..(((((((......(((((((.(((((((((.....))..)))))))...))))....(((.......)))...---.....)))......)))))))............--------- ( -13.90, z-score = 0.93, R) >consensus _UCAAUUGAAAGUAAAAGAACUGCAAUGAAUCACUGAGAUGUUUGGGAAUUAGUUAUACGGAAUUAGUAUCCCCACAAAAGUCCCCCAAAUAUUAAGUGAUUCACAAAUAUUACUCAUAG ..........................(((((((((..((((((((((............(((.......)))............)))))))))).)))))))))................ (-14.39 = -15.71 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:13 2011