| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,640,401 – 3,640,499 |
| Length | 98 |
| Max. P | 0.858253 |

| Location | 3,640,401 – 3,640,499 |
|---|---|
| Length | 98 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Shannon entropy | 0.53092 |
| G+C content | 0.57041 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.84 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
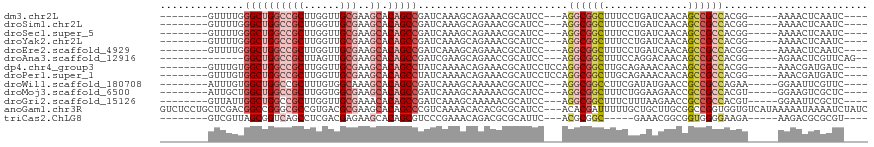
>dm3.chr2L 3640401 98 + 23011544 --------GUUUUGGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC---AGGCGGCUUUCCUGAUCAACAGCCGCCACGG-----AAAACUCAAUC---- --------((((((((((((((((......)))..)).)))))...)))))).........(((---.(((((((............)))))))..))-----)..........---- ( -34.30, z-score = -1.51, R) >droSim1.chr2L 3599803 98 + 22036055 --------GUUUUGGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC---AGGCGGCUUUCCUGAUCAACAGCCGCCACGG-----AAAACUCAAUC---- --------((((((((((((((((......)))..)).)))))...)))))).........(((---.(((((((............)))))))..))-----)..........---- ( -34.30, z-score = -1.51, R) >droSec1.super_5 1741312 98 + 5866729 --------GUUUUGGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC---AGGCGGCUUUCCUGAUCAACAGCCGCCACGG-----AAAACUCAAUC---- --------((((((((((((((((......)))..)).)))))...)))))).........(((---.(((((((............)))))))..))-----)..........---- ( -34.30, z-score = -1.51, R) >droYak2.chr2L 3634359 98 + 22324452 --------GUUUUGGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC---AGGCGGCUUUCCUGAUCAACAGCCGCCACGG-----AAAACUCAAUC---- --------((((((((((((((((......)))..)).)))))...)))))).........(((---.(((((((............)))))))..))-----)..........---- ( -34.30, z-score = -1.51, R) >droEre2.scaffold_4929 3679195 98 + 26641161 --------GUUUUGGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC---AGGCGGCUUUCCUGAUCAACAGCCGCCACGG-----AAAACUCAAUC---- --------((((((((((((((((......)))..)).)))))...)))))).........(((---.(((((((............)))))))..))-----)..........---- ( -34.30, z-score = -1.51, R) >droAna3.scaffold_12916 5695552 94 + 16180835 --------------GGCUGGCCGCUUAGUUGCGAAGCACAGCCGAUCGAAGCAGAACCGCAUCC---AGGCGGCUUUCCAGGACAACAGCCGCCACGG-----AGAACUCGUUCAG-- --------------((((((((((......)))..)).)))))((.(((.((......)).(((---.(((((((............)))))))..))-----)....))).))..-- ( -35.90, z-score = -2.22, R) >dp4.chr4_group3 6600369 101 - 11692001 --------GUUUGUGGCUGGCCGCUUGGUUGCGAAGCACAGCCUAUCAAAACAGAAACGCAUCCUCCAGGCGGCUUGCAGAAACAACAGCCGCCACGG-----AAACGAUGAUC---- --------....((((((((((((((((.((((..((...))...((......))..))))....)))))))))(((......))))))))))((((.-----...)).))...---- ( -37.60, z-score = -2.47, R) >droPer1.super_1 3699805 101 - 10282868 --------GUUUGUGGCUGGCCGCUUGGUUGCGAAGCACAGCCUAUCAAAACAGAAACGCAUCCUCCAGGCGGCUUGCAGAAACAACAGCCGCCACGG-----AAACGAUGAUC---- --------....((((((((((((((((.((((..((...))...((......))..))))....)))))))))(((......))))))))))((((.-----...)).))...---- ( -37.60, z-score = -2.47, R) >droWil1.scaffold_180708 3054346 98 - 12563649 --------AUUUGUGGCUGGCCGCUUUGUGGCAAAGCACAGCCGAUCAAAGCAAAAACGCAUCC---AGGCGGCCUUCGAUAUGAACCGCCGCCAGAA-----GGAAUUCGUUC---- --------.((((((((((...((((((...)))))).))))))..)))).....((((..(((---.((((((.(((.....)))..))))))....-----)))...)))).---- ( -31.70, z-score = -0.70, R) >droMoj3.scaffold_6500 14279693 98 - 32352404 --------AUUGCUGGCUGGCCGCUUGGUGGCGAAGCACAGCCGAUCAAAGCAAAAACGCAUCC---AGGCGGCUUUCUGGAAGAACCGCCGCCACGU-----GGAAGUCGCUC---- --------...((.((((..((((.((((((((((((...(((.......((......))....---.))).)))))..((.....))))))))).))-----)).))))))..---- ( -38.80, z-score = -1.08, R) >droGri2.scaffold_15126 4291465 98 + 8399593 --------GUUAUUGGCUGGCCGCUUGGUUGCGAAACACAGCCGAUCAAAGCAAAAACGCAUCC---AGGCGGCUUUCUUUAAGAACCGCCGCCACGU-----GGAAUUCGCUC---- --------...((((((((..(((......))).....))))))))....((......)).(((---(((((((.(((.....)))..))))))...)-----)))........---- ( -32.60, z-score = -1.26, R) >anoGam1.chr3R 20329631 115 + 53272125 GUCUCCUGCUCGACGGCCGGGCGCCGUGACGCGAAGCACAGCCCGUCAAAACACACGCGCAUCC---ACACGAUUUUUGCUGCUUGCGGCCGGUGGUGUCAUAAAAAUAAAAUCUAUC (.(.((.(((((.....)))))((((.(.(((.(((((((((.(((........))).))(((.---....)))...)).)))))))).))))))).).).................. ( -29.00, z-score = 1.67, R) >triCas2.ChLG8 5266505 93 - 15773733 --------GUCGUUAGCGGUCAGCCUCGACGAGAAGCACAGCGUCCCGAAACAGACGCGCAUUC---ACGCGGC-----GAAACGGCGGUGGGGAAGA-----AAGACGCGCGU---- --------.......((((((.((.((.....)).))...(((((........)))))...(((---.(((.((-----......)).)))..)))..-----..))).)))..---- ( -28.00, z-score = 1.63, R) >consensus ________GUUUUUGGCUGGCCGCUUGGUUGCGAAGCACAGCCGAUCAAAGCAGAAACGCAUCC___AGGCGGCUUUCCUGAACAACAGCCGCCACGG_____AAAACUCGAUC____ ..............((((((((((......)))..)).))))).........................((((((..............))))))........................ (-21.03 = -21.84 + 0.81)


| Location | 3,640,401 – 3,640,499 |
|---|---|
| Length | 98 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.19 |
| Shannon entropy | 0.53092 |
| G+C content | 0.57041 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
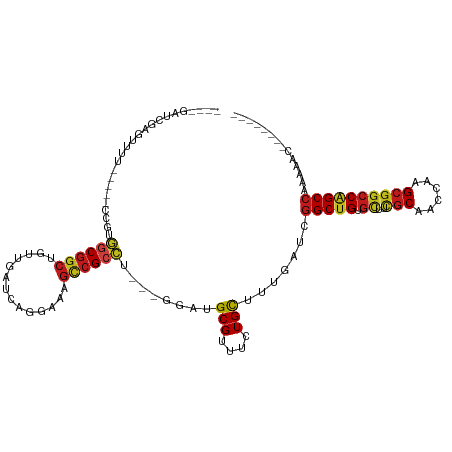
>dm3.chr2L 3640401 98 - 23011544 ----GAUUGAGUUUU-----CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU---GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCCAAAAC-------- ----(((..((...(-----(((.(((((((............))))))))---))).(((....)))))..)))(((((.((..(((......))))))))))......-------- ( -37.30, z-score = -1.62, R) >droSim1.chr2L 3599803 98 - 22036055 ----GAUUGAGUUUU-----CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU---GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCCAAAAC-------- ----(((..((...(-----(((.(((((((............))))))))---))).(((....)))))..)))(((((.((..(((......))))))))))......-------- ( -37.30, z-score = -1.62, R) >droSec1.super_5 1741312 98 - 5866729 ----GAUUGAGUUUU-----CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU---GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCCAAAAC-------- ----(((..((...(-----(((.(((((((............))))))))---))).(((....)))))..)))(((((.((..(((......))))))))))......-------- ( -37.30, z-score = -1.62, R) >droYak2.chr2L 3634359 98 - 22324452 ----GAUUGAGUUUU-----CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU---GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCCAAAAC-------- ----(((..((...(-----(((.(((((((............))))))))---))).(((....)))))..)))(((((.((..(((......))))))))))......-------- ( -37.30, z-score = -1.62, R) >droEre2.scaffold_4929 3679195 98 - 26641161 ----GAUUGAGUUUU-----CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU---GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCCAAAAC-------- ----(((..((...(-----(((.(((((((............))))))))---))).(((....)))))..)))(((((.((..(((......))))))))))......-------- ( -37.30, z-score = -1.62, R) >droAna3.scaffold_12916 5695552 94 - 16180835 --CUGAACGAGUUCU-----CCGUGGCGGCUGUUGUCCUGGAAAGCCGCCU---GGAUGCGGUUCUGCUUCGAUCGGCUGUGCUUCGCAACUAAGCGGCCAGCC-------------- --..((.((((...(-----(((.(((((((............))))))))---))).(((....))))))).))(((((.((..(((......))))))))))-------------- ( -36.90, z-score = -1.12, R) >dp4.chr4_group3 6600369 101 + 11692001 ----GAUCAUCGUUU-----CCGUGGCGGCUGUUGUUUCUGCAAGCCGCCUGGAGGAUGCGUUUCUGUUUUGAUAGGCUGUGCUUCGCAACCAAGCGGCCAGCCACAAAC-------- ----.((((.(((.(-----((..(((((((..(((....))))))))))....))).))).........)))).(((((.((..(((......))))))))))......-------- ( -37.80, z-score = -1.59, R) >droPer1.super_1 3699805 101 + 10282868 ----GAUCAUCGUUU-----CCGUGGCGGCUGUUGUUUCUGCAAGCCGCCUGGAGGAUGCGUUUCUGUUUUGAUAGGCUGUGCUUCGCAACCAAGCGGCCAGCCACAAAC-------- ----.((((.(((.(-----((..(((((((..(((....))))))))))....))).))).........)))).(((((.((..(((......))))))))))......-------- ( -37.80, z-score = -1.59, R) >droWil1.scaffold_180708 3054346 98 + 12563649 ----GAACGAAUUCC-----UUCUGGCGGCGGUUCAUAUCGAAGGCCGCCU---GGAUGCGUUUUUGCUUUGAUCGGCUGUGCUUUGCCACAAAGCGGCCAGCCACAAAU-------- ----(((((...(((-----....((((((..(((.....))).)))))).---)))..)))))...........((((((((((((...)))))))..)))))......-------- ( -35.70, z-score = -1.44, R) >droMoj3.scaffold_6500 14279693 98 + 32352404 ----GAGCGACUUCC-----ACGUGGCGGCGGUUCUUCCAGAAAGCCGCCU---GGAUGCGUUUUUGCUUUGAUCGGCUGUGCUUCGCCACCAAGCGGCCAGCCAGCAAU-------- ----(((((...(((-----(...((((((..(((.....))).)))))))---)))..)))))(((((......(((((.((..(((......))))))))))))))).-------- ( -39.60, z-score = -1.41, R) >droGri2.scaffold_15126 4291465 98 - 8399593 ----GAGCGAAUUCC-----ACGUGGCGGCGGUUCUUAAAGAAAGCCGCCU---GGAUGCGUUUUUGCUUUGAUCGGCUGUGUUUCGCAACCAAGCGGCCAGCCAAUAAC-------- ----(((((((.(((-----(...((((((..(((.....))).)))))))---)))......))))))).....(((((.((..(((......))))))))))......-------- ( -36.90, z-score = -1.83, R) >anoGam1.chr3R 20329631 115 - 53272125 GAUAGAUUUUAUUUUUAUGACACCACCGGCCGCAAGCAGCAAAAAUCGUGU---GGAUGCGCGUGUGUUUUGACGGGCUGUGCUUCGCGUCACGGCGCCCGGCCGUCGAGCAGGAGAC .(((((.......))))).....(((.(.(((((((..(((.....(((((---....)))))..))))))).))).).)))((((((.(((((((.....))))).)))).)))).. ( -32.30, z-score = 1.89, R) >triCas2.ChLG8 5266505 93 + 15773733 ----ACGCGCGUCUU-----UCUUCCCCACCGCCGUUUC-----GCCGCGU---GAAUGCGCGUCUGUUUCGGGACGCUGUGCUUCUCGUCGAGGCUGACCGCUAACGAC-------- ----..(((.(((..-----........((.((.((..(-----(.(((((---....))))).......))..)))).))(((((.....))))).)))))).......-------- ( -26.40, z-score = 1.54, R) >consensus ____GAUCGAGUUUU_____CCGUGGCGGCUGUUGAUCAGGAAAGCCGCCU___GGAUGCGUUUCUGCUUUGAUCGGCUGUGCUUCGCAACCAAGCGGCCAGCCAAAAAC________ ........................((((((..............))))))........(((....))).......(((((.((..(((......)))))))))).............. (-21.39 = -21.72 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:20 2011