| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,261,420 – 18,261,483 |
| Length | 63 |
| Max. P | 0.526036 |

| Location | 18,261,420 – 18,261,483 |
|---|---|
| Length | 63 |
| Sequences | 10 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.50474 |
| G+C content | 0.43853 |
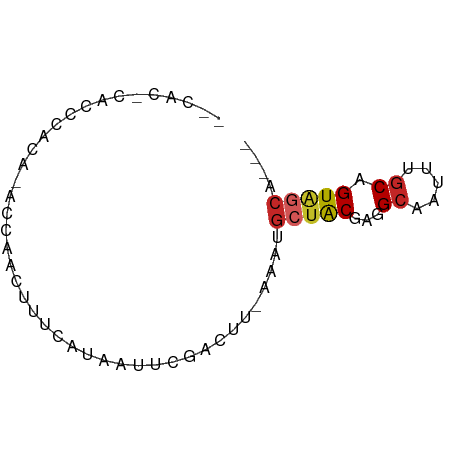
| Mean single sequence MFE | -11.23 |
| Consensus MFE | -7.15 |
| Energy contribution | -7.33 |
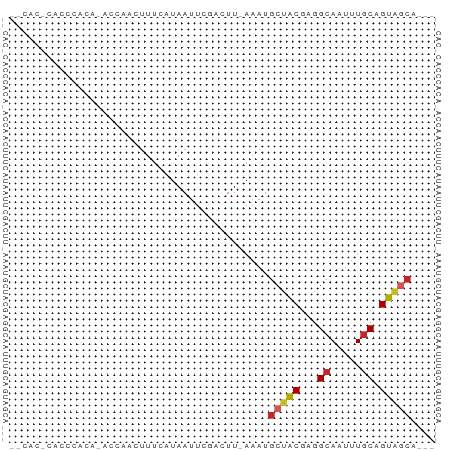
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.526036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
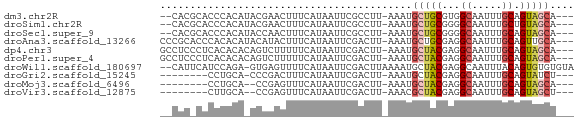
>dm3.chr2R 18261420 63 - 21146708 --CACGCACCCACAUACGAACUUUCAUAAUUCGCCUU-AAAUGCUGCGUGGCAAUUUGCAGUAGCA--- --...((.........((((.........))))....-...(((((((........))))))))).--- ( -11.60, z-score = -0.17, R) >droSim1.chr2R 16858898 63 - 19596830 --CACGCACCCACAUACGAACUUUCAUAAUUCGCCUU-AAAUGCUGCGGGGCAAUUUGCUGUAGCA--- --..............((((.........))))....-...((((((((.(.....).))))))))--- ( -10.30, z-score = 0.75, R) >droSec1.super_9 1569505 63 - 3197100 --CACGCACCCACAUACCAACUUUCAUAAUUCGCCUU-AAAUGCUGCGGGGCAAUUUGCAGUAGCA--- --...................................-...((((((...((.....)).))))))--- ( -10.00, z-score = 0.69, R) >droAna3.scaffold_13266 12389142 65 + 19884421 CCCGCACCCACACAUACAUACUUUCAUAAUUCGACUU-AAAUGCUGCGAGGCAAUUUGCAGUUGCA--- ...(((.................((.......))...-....((((((((....))))))))))).--- ( -9.80, z-score = -0.29, R) >dp4.chr3 4906846 65 + 19779522 GCCUCCCUCACACACAGUCUUUUUCAUAAUUCGACUU-AAAUGCUACGAGGCAAUUUGCAGUAGCA--- (((((..........((((.............)))).-.........)))))....(((....)))--- ( -11.37, z-score = -1.54, R) >droPer1.super_4 4179292 65 - 7162766 GCCUCCCUCACACACAGUCUUUUUCAUAAUUCGACUU-AAAUGCUACGAGGCAAUUUGCAGUAGCA--- (((((..........((((.............)))).-.........)))))....(((....)))--- ( -11.37, z-score = -1.54, R) >droWil1.scaffold_180697 4051158 66 + 4168966 --CAUUCAUCCAGA-GUGAGUUUUCAUAAUUCGACUUAAAAUGCUACGAGGCAAUUUACAGUGUGUGUA --(((.(((...((-(((((((.....))))).))))....((((....)))).......))).))).. ( -9.80, z-score = 0.35, R) >droGri2.scaffold_15245 13306236 56 - 18325388 --------CCUGCA-CCCGACUUUCAUAAUUCGACUU-AAAUGCUACGAGGCAAUUUGCAGUAUCU--- --------.(((((-..(((.((....)).)))....-...((((....))))...))))).....--- ( -9.90, z-score = -1.50, R) >droMoj3.scaffold_6496 12955488 55 - 26866924 --------CCUGCA--CCGAGUUUCAUAAUUCGACUU-AAAUGCUACGAGGCAAUUUGCAGUAGCA--- --------......--.((((((....))))))....-...((((((...((.....)).))))))--- ( -14.30, z-score = -2.15, R) >droVir3.scaffold_12875 19264786 55 - 20611582 --------CUUGCA--CCGAGUUUCAUAAUUCGACUU-AAACGCUACGAGGCAAUUUGCAGUAGCU--- --------......--.((((((....))))))....-....(((((...((.....)).))))).--- ( -13.90, z-score = -1.98, R) >consensus __CAC_CACCCACA_ACCAACUUUCAUAAUUCGACUU_AAAUGCUACGAGGCAAUUUGCAGUAGCA___ ..........................................(((((...((.....)).))))).... ( -7.15 = -7.33 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:05 2011