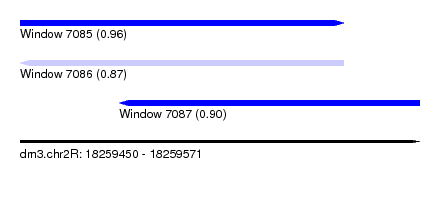
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,259,450 – 18,259,571 |
| Length | 121 |
| Max. P | 0.956626 |

| Location | 18,259,450 – 18,259,548 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Shannon entropy | 0.47189 |
| G+C content | 0.39023 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.83 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
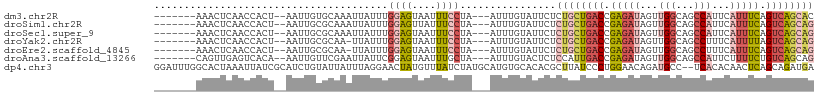
>dm3.chr2R 18259450 98 + 21146708 -------AAACUCAACCACU--AAUUGUGCAAAUUAUUUGGAGUAAUUUCCUA---AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAC -------.............--....((((((((((...((((....))))))---))))))))....(((((((.(((((...(((...)))...))))).))))))). ( -26.40, z-score = -3.31, R) >droSim1.chr2R 16856979 98 + 19596830 -------AAACUCAACCACU--AAUUGCGCAAAUUAUUUGGAGUUAUUUCCUA---AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAG -------.............--......((((((((...((((....))))))---)))))).....((((((((.(((((...(((...)))...))))).)))))))) ( -26.80, z-score = -3.54, R) >droSec1.super_9 1567471 98 + 3197100 -------AAACUCAACCACU--AAUUGCGCAAAUUAUUUGGAGUAAUUUCCUA---AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAG -------.............--......((((((((...((((....))))))---)))))).....((((((((.(((((...(((...)))...))))).)))))))) ( -26.80, z-score = -3.51, R) >droYak2.chr2R 18008057 97 + 21139217 -------AAACUCAACCACU--AAUUGCGCAA-UUAUUUGGAGUAAUUUCCUA---AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUUAGUCAGCAG -------.............--......((((-......((((....))))..---..)))).....((((((((..((((.(..((...))..).))))..)))))))) ( -21.20, z-score = -1.60, R) >droEre2.scaffold_4845 12399490 97 + 22589142 -------AAACUCAACCACU--AAUUGCGCAA-UUAUUUGGAGUAAUUUCCUA---AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUCAGUCAGCAG -------.............--......((((-......((((....))))..---..)))).....((((((((.(((((.(..((...))..).))))).)))))))) ( -22.30, z-score = -1.93, R) >droAna3.scaffold_13266 12386568 98 - 19884421 -------CAGUUGAGUCACA--AAUUGUUCGAAUUAUUCGGAGUAAUUUGCUA---AUUUGUACUCUCCAUUGACCGAGAUAGUUGGCAGCCAUUCUUUUCUGUCAGCAG -------((((.((((.(((--(((((..((((((((.....)))))))).))---))))))))))...)))).........((((((((..........)))))))).. ( -25.40, z-score = -2.25, R) >dp4.chr3 4905245 108 - 19779522 GGAUUUGGCACUAAAUUAUCGCAUCUGUAUUAUUUAGGAACUAUGUUUAUCUAUGCAUGUGCACACGCUUAUCCCUGGAACAGAUGCC--UCACACAACUCAGCAGAUGA .((((((....))))))...((((((((....((((((...(((((........))))).((....)).....)))))))))))))).--.................... ( -20.80, z-score = 0.26, R) >consensus _______AAACUCAACCACU__AAUUGCGCAAAUUAUUUGGAGUAAUUUCCUA___AUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAG ............................((((.......((((....)))).......)))).....((((((((.(((((...((.....))...))))).)))))))) (-11.98 = -13.83 + 1.84)
| Location | 18,259,450 – 18,259,548 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.34 |
| Shannon entropy | 0.47189 |
| G+C content | 0.39023 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -13.32 |
| Energy contribution | -14.33 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
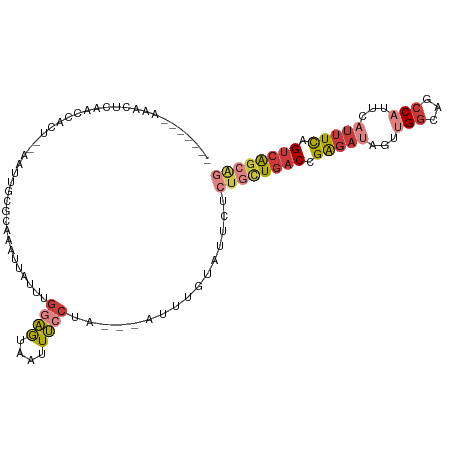
>dm3.chr2R 18259450 98 - 21146708 GUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU---UAGGAAAUUACUCCAAAUAAUUUGCACAAUU--AGUGGUUGAGUUU------- .((((((((((.(((..(((...)))..))).)))))))))).......(((((---(((((......)))...)))))))(((....--.))).........------- ( -25.10, z-score = -1.99, R) >droSim1.chr2R 16856979 98 - 19596830 CUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU---UAGGAAAUAACUCCAAAUAAUUUGCGCAAUU--AGUGGUUGAGUUU------- (((((((((((.(((..(((...)))..))).)))))))))))......(((((---(((((......)))...)))))))(((....--.))).........------- ( -27.40, z-score = -2.78, R) >droSec1.super_9 1567471 98 - 3197100 CUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU---UAGGAAAUUACUCCAAAUAAUUUGCGCAAUU--AGUGGUUGAGUUU------- (((((((((((.(((..(((...)))..))).)))))))))))......(((((---(((((......)))...)))))))(((....--.))).........------- ( -27.40, z-score = -2.67, R) >droYak2.chr2R 18008057 97 - 21139217 CUGCUGACUAAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU---UAGGAAAUUACUCCAAAUAA-UUGCGCAAUU--AGUGGUUGAGUUU------- (((((((((.....((.((........)).))..)))))))))...........---..(((......)))...(((-(..(......--.)..)))).....------- ( -22.30, z-score = -1.31, R) >droEre2.scaffold_4845 12399490 97 - 22589142 CUGCUGACUGAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU---UAGGAAAUUACUCCAAAUAA-UUGCGCAAUU--AGUGGUUGAGUUU------- (((((((((((...((.((........)).)))))))))))))...........---..(((......)))...(((-(..(......--.)..)))).....------- ( -26.10, z-score = -2.32, R) >droAna3.scaffold_13266 12386568 98 + 19884421 CUGCUGACAGAAAAGAAUGGCUGCCAACUAUCUCGGUCAAUGGAGAGUACAAAU---UAGCAAAUUACUCCGAAUAAUUCGAACAAUU--UGUGACUCAACUG------- ((..((((.((......(((...)))......)).))))..)).((((((((((---(..(.(((((.......))))).)...))))--))).)))).....------- ( -16.00, z-score = 0.09, R) >dp4.chr3 4905245 108 + 19779522 UCAUCUGCUGAGUUGUGUGA--GGCAUCUGUUCCAGGGAUAAGCGUGUGCACAUGCAUAGAUAAACAUAGUUCCUAAAUAAUACAGAUGCGAUAAUUUAGUGCCAAAUCC ......((((((((((....--.((((((((...((((((..(((((....)))))...(.....)...)))))).......)))))))).))))))))))......... ( -27.50, z-score = -1.12, R) >consensus CUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAU___UAGGAAAUUACUCCAAAUAAUUUGCGCAAUU__AGUGGUUGAGUUU_______ (((((((((((.(((..((.....))..))).)))))))))))................(((......)))..........(((.......)))................ (-13.32 = -14.33 + 1.01)

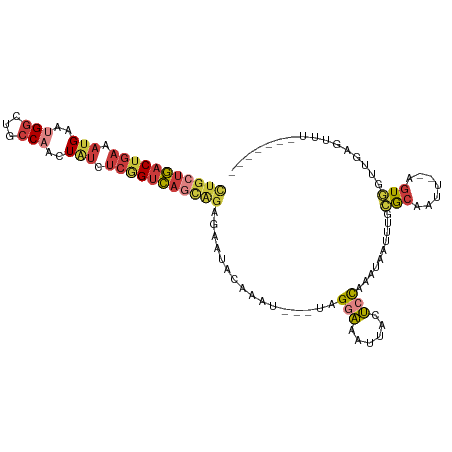
| Location | 18,259,480 – 18,259,571 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 93.21 |
| Shannon entropy | 0.13009 |
| G+C content | 0.42417 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -21.57 |
| Energy contribution | -22.98 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18259480 91 - 21146708 AUAUUAAUUGGCCUGGUUCGG-UGGUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCC ....(((((..(((((((..(-((.((((((((((.(((..(((...)))..))).)))))))))).....))).))))))).))))).... ( -26.60, z-score = -2.33, R) >droSim1.chr2R 16857009 91 - 19596830 AUAUUAAUUGGCCUGGUUCGG-UGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUAACUCC ...........(((((((..(-(.(((((((((((.(((..(((...)))..))).)))))))))))...))...))))))).......... ( -27.90, z-score = -3.04, R) >droSec1.super_9 1567501 91 - 3197100 AUAUUAAUUGGCCUGGUUCGG-CGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCC ....(((((..(((((((.(.-(.(((((((((((.(((..(((...)))..))).))))))))))).)....).))))))).))))).... ( -30.10, z-score = -3.55, R) >droYak2.chr2R 18008086 91 - 21139217 AUAUUAAUUGGCCUGGUUCGG-UGCUGCUGACUAAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCC ....(((((..(((((((..(-(.(((((((((.....((.((........)).))..)))))))))...))...))))))).))))).... ( -24.40, z-score = -1.92, R) >droEre2.scaffold_4845 12399519 91 - 22589142 AUAUUAAUUGGCCUGGUUCGG-UGCUGCUGACUGAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCC ....(((((..(((((((..(-(.(((((((((((...((.((........)).)))))))))))))...))...))))))).))))).... ( -28.20, z-score = -2.96, R) >droAna3.scaffold_13266 12386598 92 + 19884421 AUAUUAAUUGGUCCUAUUUGGCUGCUGCUGACAGAAAAGAAUGGCUGCCAACUAUCUCGGUCAAUGGAGAGUACAAAUUAGCAAAUUACUCC ......((((..(.((.(((((.(((.((........))...))).))))).))....)..))))...(((((.............))))). ( -17.12, z-score = 0.47, R) >consensus AUAUUAAUUGGCCUGGUUCGG_UGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCC ....(((((..(((((((.(....(((((((((((.(((..(((...)))..))).)))))))))))......).))))))).))))).... (-21.57 = -22.98 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:04 2011