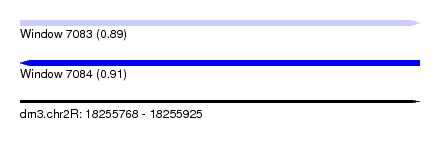
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,255,768 – 18,255,925 |
| Length | 157 |
| Max. P | 0.908123 |

| Location | 18,255,768 – 18,255,925 |
|---|---|
| Length | 157 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | forward |
| Mean pairwise identity | 86.10 |
| Shannon entropy | 0.26241 |
| G+C content | 0.41113 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -27.82 |
| Energy contribution | -28.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.886523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
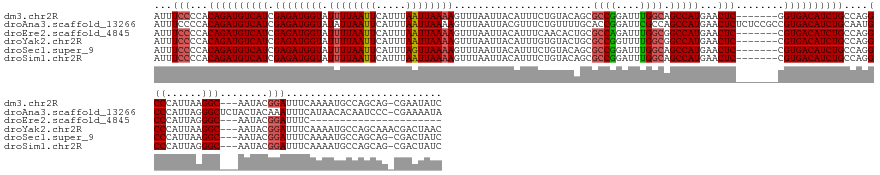
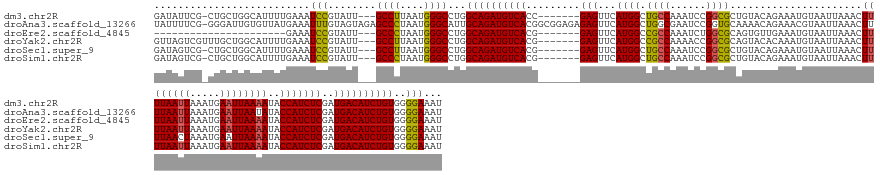
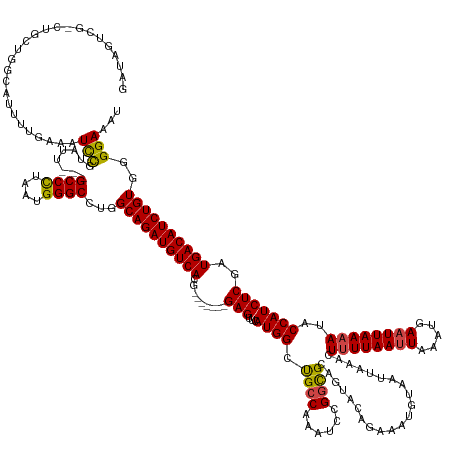
>dm3.chr2R 18255768 157 + 21146708 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAAUUAAAAGUUUAAUUACAUUUCUGUACAGCGCCGGAUUUGGCAGCCAUGAACUC-------GGUGACAUCUGCCAGGCCCAUUAAGGC---AAUACGGAUUUCAAAAUGCCAGCAG-CGAAUAUC ...(((...(((((((((((((((((((.((((((((.....))))))))(((....((((....)))).)))((((....)))).)))))...)))-------)))))))))))....(((......)))---.....)))........(((.....)-))...... ( -42.30, z-score = -2.46, R) >droAna3.scaffold_13266 12383026 167 - 19884421 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUAUUAAUUCAUUUAAUUAAAAGUUUAAUUACGUUUCUGUUUUGCACCGGAUUCGCCAGCCAUGAACUCUCUCCGCCGUGACAUCUGCAAUGCCCAUUAGGGCUCUACUACAAAUUUCAUAACACAAUCCC-CGAAAAUA .((((....((((((((((.((((.(((.............(((((((....)))))))((..((((........))))..))..........))).))))....))))))))))....((((....))))............................-.))))... ( -34.50, z-score = -2.03, R) >droEre2.scaffold_4845 12395756 136 + 22589142 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAAUUAAAAGUUUAAUUACAUUUCAACACUGCGCCAGAUUUGGCGGCCAUGAACUC-------CGUGACAUCUGCCAGGCCCAUUAGGGC---AAUACGGAUUUC---------------------- ...(((...((((((((((.((((((((.((((((((.....))))))))......................((((......)))))))))...)))-------.))))))))))....((((....))))---.....)))....---------------------- ( -42.50, z-score = -4.34, R) >droYak2.chr2R 18003834 158 + 21139217 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAAUUAAAAGUUUAAUUACAUUUGUGUACUGCGCCGGUUUUGGCGGCCAUGAACUC-------CGUGACAUCUGCCAGGCCCAUUAAGGC---AAUACGGAUUUCAAAAUGCCAGCAAACGACUAAC ...(((...((((((((((.((((((((.((((((((.....)))))))).......((((....))))...(((((....))))))))))...)))-------.))))))))))....(((......)))---.....))).......................... ( -38.70, z-score = -1.12, R) >droSec1.super_9 1563735 157 + 3197100 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAGUUAAAAGUUUAAUUACAUUUCUGUACAGCGCCGGAUUUGGCAGCCAUGAACUC-------CGUGACAUCUGCCAGGCCCAUUAAGGC---AAUACGGAUUUCAAAAUGCCAGCAG-CGACUAUC ...(((...((((((((((.((((((((.((((((((.....))))))))(((....((((....)))).)))((((....)))).)))))...)))-------.))))))))))....(((......)))---.....)))........(((.....)-))...... ( -37.30, z-score = -1.20, R) >droSim1.chr2R 16853225 157 + 19596830 AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAAUUAAAAGUUUAAUUACAUUUCUGUACAGCGCCGGAUUUGGCAGCCAUGAACUC-------CGUGACAUCUGCCAGGCCCAUUAGGGC---AAUACGGAUUUCAAAAUGCCAGCAG-CGACUAUC ...(((...((((((((((.((((((((.((((((((.....))))))))(((....((((....)))).)))((((....)))).)))))...)))-------.))))))))))....((((....))))---.....)))........(((.....)-))...... ( -40.90, z-score = -1.97, R) >consensus AUUUCCCCACAGAUGUCAUCGAGAUGGUAUUUUAAUUCAUUUAAUUAAAAGUUUAAUUACAUUUCUGUACAGCGCCGGAUUUGGCAGCCAUGAACUC_______CGUGACAUCUGCCAGGCCCAUUAAGGC___AAUACGGAUUUCAAAAUGCCAGCAG_CGACUAUC ...(((...((((((((((.((((((...((((((((.....)))))))).........))))))......((((((....)))).)).................))))))))))....(((......)))........))).......................... (-27.82 = -28.27 + 0.45)
| Location | 18,255,768 – 18,255,925 |
|---|---|
| Length | 157 |
| Sequences | 6 |
| Columns | 168 |
| Reading direction | reverse |
| Mean pairwise identity | 86.10 |
| Shannon entropy | 0.26241 |
| G+C content | 0.41113 |
| Mean single sequence MFE | -44.12 |
| Consensus MFE | -34.65 |
| Energy contribution | -34.48 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
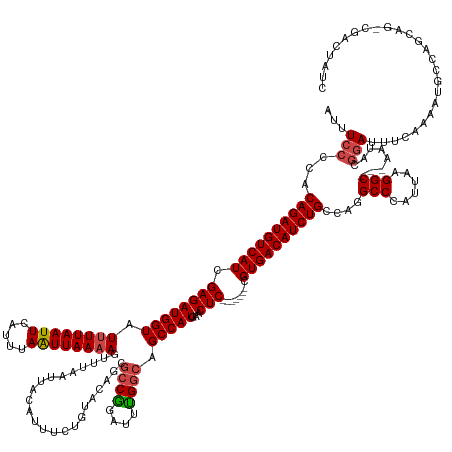
>dm3.chr2R 18255768 157 - 21146708 GAUAUUCG-CUGCUGGCAUUUUGAAAUCCGUAUU---GCCUUAAUGGGCCUGGCAGAUGUCACC-------GAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU ........-.....((((...((.....))...)---)))........(((.((((((((((.(-------(((...((((..(((......)))....((((....)))).......((((((((.....))))))))..)))))))).)))))))))).))).... ( -46.10, z-score = -2.57, R) >droAna3.scaffold_13266 12383026 167 + 19884421 UAUUUUCG-GGGAUUGUGUUAUGAAAUUUGUAGUAGAGCCCUAAUGGGCAUUGCAGAUGUCACGGCGGAGAGAGUUCAUGGCUGGCGAAUCCGGUGCAAAACAGAAACGUAAUUAAACUUUUAAUUAAAUGAAUUAAUAUACCAUCUCGAUGACAUCUGUGGGGAAAU ..((..((-.((((.((((..(((.(((((((((...((((....))))))))))))).)))..)).((((.(((((((.(((((.....)))))..............(((((((....))))))).))))))).........))))....)))))).))..))... ( -44.60, z-score = -1.55, R) >droEre2.scaffold_4845 12395756 136 - 22589142 ----------------------GAAAUCCGUAUU---GCCCUAAUGGGCCUGGCAGAUGUCACG-------GAGUUCAUGGCCGCCAAAUCUGGCGCAGUGUUGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU ----------------------....(((.....---((((....)))).(.((((((((((..-------(((...((((.(((((....)))))((..(((((((.((......)))))))))....))..........)))))))..)))))))))).))))... ( -42.10, z-score = -2.94, R) >droYak2.chr2R 18003834 158 - 21139217 GUUAGUCGUUUGCUGGCAUUUUGAAAUCCGUAUU---GCCUUAAUGGGCCUGGCAGAUGUCACG-------GAGUUCAUGGCCGCCAAAACCGGCGCAGUACACAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU .....(((((....((((...((.....))...)---)))..))))).(((.((((((((((..-------(((...((((.((((......))))...((((....)))).......((((((((.....))))))))..)))))))..)))))))))).))).... ( -43.90, z-score = -1.52, R) >droSec1.super_9 1563735 157 - 3197100 GAUAGUCG-CUGCUGGCAUUUUGAAAUCCGUAUU---GCCUUAAUGGGCCUGGCAGAUGUCACG-------GAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAACUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU ........-.....((((...((.....))...)---)))........(((.((((((((((((-------(((((((((((.(((......)))))).((((....)))).................)))))))...........))).)))))))))).))).... ( -43.00, z-score = -1.42, R) >droSim1.chr2R 16853225 157 - 19596830 GAUAGUCG-CUGCUGGCAUUUUGAAAUCCGUAUU---GCCCUAAUGGGCCUGGCAGAUGUCACG-------GAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU ......((-(((.((((((..((((.(((((...---((((....))))..(((....))))))-------)).))))..).)))))....)))))((.((((((..(((........((((((((.....))))))))...(((....)))))))))))).)).... ( -45.00, z-score = -1.79, R) >consensus GAUAGUCG_CUGCUGGCAUUUUGAAAUCCGUAUU___GCCCUAAUGGGCCUGGCAGAUGUCACG_______GAGUUCAUGGCUGCCAAAUCCGGCGCAGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCGAUGACAUCUGUGGGGAAAU ..........................(((........((((....))))...((((((((((((........(((((((.((.(((......)))))............(((((((....))))))).)))))))............)).))))))))))..)))... (-34.65 = -34.48 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:43:02 2011