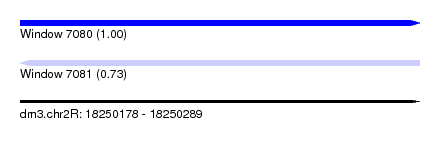
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,250,178 – 18,250,289 |
| Length | 111 |
| Max. P | 0.995667 |

| Location | 18,250,178 – 18,250,289 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.49577 |
| G+C content | 0.37246 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.38 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
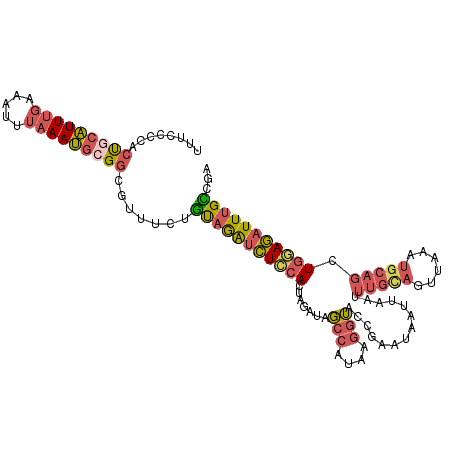
>dm3.chr2R 18250178 111 + 21146708 UUUCCCCACUCCAUUUGUAAUUUAAAUGCGGCGUUUCUGUAGAUCUCCACUAGAUAGCCAUAAGGUACCGAAUAAUUAAUUUGUAGUUAAAUGCAGCUGGAGAUUUGCCGA ......(.((.((((((.....)))))).)).)..((.(((((((((((.......(((....)))..............(((((......))))).))))))))))).)) ( -24.10, z-score = -1.18, R) >droYak2.chr2R 17998023 111 + 21139217 UUUCGCCACUGCAUUUGCAAUUUAAAUGCGGCAUUUCUGUAGAUCUUCACUUGAUAGCCAUAAGGUACCCAAUAAUUAAUUUGCAGUUAAAUGCAGCUGCAGUUUUGUCGA ..((((.((((((.(((((.(((((.(((((((((...((.(.....)))..))).)))....((...))............))).)))))))))).))))))...).))) ( -25.60, z-score = -0.88, R) >droSec1.super_9 1558124 111 + 3197100 UUUCCCCGCUGCAUUUGUAAUUUAAAUGCGGCGUUUCUGUAGAUCUCCACUAGAUAGCCAUAAGGUGCCGAAUAAUUAAUUUGCAGUUAAAUGCAGCUGGAGAUUUGCCGA ......(((((((((((.....)))))))))))..((.(((((((((((.......(((....)))..............(((((......))))).))))))))))).)) ( -37.40, z-score = -4.07, R) >droSim1.chr2R 16847511 111 + 19596830 UUUUCCCGCUGCAUUUGAAAUUUAAAUGCGGCGUUUCUGUAGAUCUCCACUAGAUAGCCAUAAGGUACCGAAUAAAUAAUUUGCAGUUAAAUGCAGCUGGAGAUUUGCCGA ......((((((((((((...))))))))))))..((.(((((((((((.......(((....)))..............(((((......))))).))))))))))).)) ( -37.00, z-score = -4.39, R) >droWil1.scaffold_180697 4034030 105 - 4168966 -UUUAACAGCAUUUUAAAAGUUGAAAGCCGGAAAUACAUCAAGUUUUUA----ACAUCGGUUGGAAAUUGACAAAACACCACA-AGUAAAAGAAUGCUUAAAAUUAGAAUA -......((((((((....(((((((((..((......))..)))))))----))...(((((........))....)))...-......))))))))............. ( -18.40, z-score = -2.11, R) >consensus UUUCCCCACUGCAUUUGAAAUUUAAAUGCGGCGUUUCUGUAGAUCUCCACUAGAUAGCCAUAAGGUACCGAAUAAUUAAUUUGCAGUUAAAUGCAGCUGGAGAUUUGCCGA ........(((((((((.....))))))))).......(((((((((((.......(((....)))..............(((((......))))).)))))))))))... (-16.38 = -17.38 + 1.00)

| Location | 18,250,178 – 18,250,289 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.61 |
| Shannon entropy | 0.49577 |
| G+C content | 0.37246 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.18 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
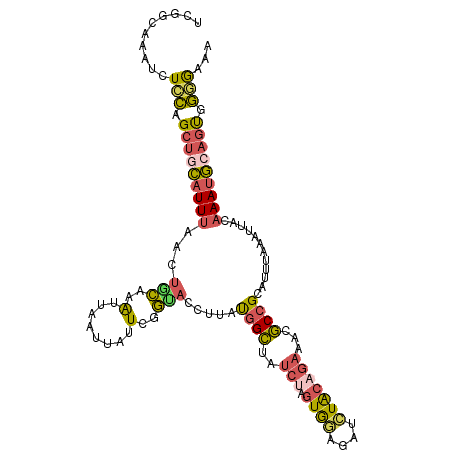
>dm3.chr2R 18250178 111 - 21146708 UCGGCAAAUCUCCAGCUGCAUUUAACUACAAAUUAAUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGGAGUGGGGAAA ........((((((.((.(((((...................((((((....)).))))..((((...((....))....))))...........))))).)))))))).. ( -23.50, z-score = -0.74, R) >droYak2.chr2R 17998023 111 - 21139217 UCGACAAAACUGCAGCUGCAUUUAACUGCAAAUUAAUUAUUGGGUACCUUAUGGCUAUCAAGUGAAGAUCUACAGAAAUGCCGCAUUUAAAUUGCAAAUGCAGUGGCGAAA (((.(...((((((..(((((((((.(((.......(((((.((((((....)).)))).))))).......((....))..))).))))).))))..)))))).)))).. ( -30.70, z-score = -2.45, R) >droSec1.super_9 1558124 111 - 3197100 UCGGCAAAUCUCCAGCUGCAUUUAACUGCAAAUUAAUUAUUCGGCACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGCAGCGGGGAAA ........(((((.((((((((....((.((.((((.....((((.((....))...(((.((((....)))))))...))))...)))).)).))))))))))))))).. ( -31.30, z-score = -2.65, R) >droSim1.chr2R 16847511 111 - 19596830 UCGGCAAAUCUCCAGCUGCAUUUAACUGCAAAUUAUUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUUCAAAUGCAGCGGGAAAA ..........(((.(((((((((.((((.((........)))))).......(((..(((.((((....)))))))...))).............))))))))).)))... ( -28.00, z-score = -1.87, R) >droWil1.scaffold_180697 4034030 105 + 4168966 UAUUCUAAUUUUAAGCAUUCUUUUACU-UGUGGUGUUUUGUCAAUUUCCAACCGAUGU----UAAAAACUUGAUGUAUUUCCGGCUUUCAACUUUUAAAAUGCUGUUAAA- .............((((((........-..((((................))))..((----(.(((.((.((......)).)).))).)))......))))))......- ( -9.59, z-score = 1.23, R) >consensus UCGGCAAAUCUCCAGCUGCAUUUAACUGCAAAUUAAUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGCAGUGGGGAAA ..........(((.(((((((((...(((..(........)..))).....((((......((((....))))......))))............))))))))).)))... (-13.90 = -13.18 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:59 2011