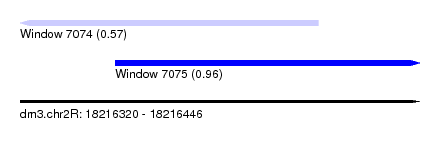
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,216,320 – 18,216,446 |
| Length | 126 |
| Max. P | 0.960470 |

| Location | 18,216,320 – 18,216,414 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.58 |
| Shannon entropy | 0.75942 |
| G+C content | 0.48765 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -10.36 |
| Energy contribution | -9.76 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.569955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18216320 94 - 21146708 GAAACGGUUGCUCAUCAAACGCGUUC----UCUGAAAG------CCCACCUCUGCUGUUGCCAAGGACCUUUGA-CUGGUCUUAACAGUAUUUGAUGGUACAGGA--------- ........(((.(((((((.((.(((----...))).)------).......((((((((....(((((.....-..))))))))))))))))))))))).....--------- ( -22.90, z-score = -0.08, R) >droAna3.scaffold_13266 12343508 82 + 19884421 GAAACGGUUGCCAAUCGAACUCCGACCCAAUC-CGA------------------CCUCUGCAGUUGCCAAAAG--AUUG--GCAACAGGAUUAGGCCAGAUGAGC--------- .........(((..(((.....)))...((((-((.------------------...)....((((((((...--.)))--))))).))))).))).........--------- ( -20.80, z-score = -0.98, R) >droEre2.scaffold_4845 12356256 85 - 22589142 GAAACGGUUGAACAUCAAACGCGUUC----UCUGAAAA------C---------CUGUUGCCAAGGACCUUUGA-CUGGUCUUAACAGGAUUAGGUGGUACGUGC--------- ...................(((((((----(((((...------(---------((((((....(((((.....-..)))))))))))).))))).)).))))).--------- ( -22.30, z-score = -0.82, R) >droYak2.chr2R 17967620 94 - 21139217 GAAACGGUUGAUCAUCAAACGCGUUC----UUUGAAAA------CCCAUCUCUUCUGUUACCAAGGAUCUUUGA-GUGGUCCUAACAUGAUUAGGUGGUACAGGC--------- ..(((((((........))).)))).----........------.((((((..((((((....((((((.....-..)))))))))).))..)))))).......--------- ( -21.80, z-score = -0.25, R) >droSec1.super_9 1526646 94 - 3197100 GAAACGGUUGCUAAUCAAACGCGUUC----UCUGAAAA------CCCACCUCUGCUGUUGCCAAGGACCUUUGA-CUGGUCUUAACAGUAUUAGAUGGUACAGGA--------- (((((((((........))).)))).----))......------((.(((((((((((((....(((((.....-..))))))))))))...))).)))...)).--------- ( -22.50, z-score = -0.53, R) >droSim1.chr2R 16815676 94 - 19596830 GAAACGGUUGCUAAUCAAACGCGUUC----UCUGAAAA------CCCACCUCUGCUGUUGCCAAGGACCUUUGA-CUGGUCUUAACAGUAUUAGAUGGUACAGGA--------- (((((((((........))).)))).----))......------((.(((((((((((((....(((((.....-..))))))))))))...))).)))...)).--------- ( -22.50, z-score = -0.53, R) >droPer1.super_4 4136447 114 - 7162766 GAAACGGCUGCUAAUCGAGCUCGGUCUCCGUCCGGGAAUGUUGCCCUGUUUCUGCUGUUGUCCCCCACCUGGGAGGCGGUGGCAACAGGAUUAAGUACAACAUACAUAUGCAGC ......(((((........(((((.......))))).((((((.(((((...(((..(((((.(((....))).)))))..))))))))........))))))......))))) ( -39.80, z-score = -1.09, R) >dp4.chr3 4864026 114 + 19779522 GAAACGGCUGCUAAUCGAGCUCGGUCUCCGUCCGGGAGUGUUGCCCUGUUUCUGCUGUUGUUCCCCACCUGGGAGGCGGUGGCAACAGGAUUAAGUACAACAUACAUAUGCAGC ......(((((........(((((.......))))).((((((.(((((...(((..((((..(((....)))..))))..))))))))........))))))......))))) ( -39.90, z-score = -0.91, R) >droGri2.scaffold_15245 13245942 85 - 18325388 GAAACGGUUGCUAAUUG--UUCGCACACGAUCUCAA------------------CUUCUGCCUUUUGCCAUCG--GUGGCUGCAACCGAAUUAAGAACAACAACAAC------- ....(((((((...(((--.(((....)))...)))------------------............((((...--.)))).)))))))...................------- ( -19.10, z-score = -1.44, R) >consensus GAAACGGUUGCUAAUCAAACGCGUUC____UCUGAAAA______CCCA_CUCUGCUGUUGCCAAGGACCUUUGA_CUGGUCGUAACAGGAUUAGGUGGUACAGGC_________ ....(((((........))).))...............................((((((....(((((........))))))))))).......................... (-10.36 = -9.76 + -0.60)


| Location | 18,216,350 – 18,216,446 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.80 |
| Shannon entropy | 0.68400 |
| G+C content | 0.49307 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18216350 96 + 21146708 UCAAAGGUCCUUGGCAACAGCAGAGGUGGG------CUUUCAGA----GAACGCGUUUGAUGAGCAACCGUUUCGAAAGCAACGGAUUUGCAAUUUAUCAAACCUC ..........(((....)))..((((((..------(((...))----)..)))((((((((((((((((((.(....).)))))..))))...)))))))))))) ( -28.20, z-score = -1.69, R) >droAna3.scaffold_13266 12343535 84 - 19884421 ------UCUUUUGGCAACUGCAGAGG-------------UCGGAUUGGGUCGGAGUUCGAUUGGCAACCGUUUCGAGAGGAACGGUCCUGCCCCUGAUUGAAC--- ------.((((((.(....)))))))-------------((.(((...))).))((((((((((((((((((((....))))))))..))))...))))))))--- ( -29.60, z-score = -1.05, R) >droEre2.scaffold_4845 12356286 87 + 22589142 UCAAAGGUCCUUGGCAACAG---------G------UUUUCAGA----GAACGCGUUUGAUGUUCAACCGUUUCGAAAGCAACGGACUUGCAAUUUAUCGAACCUC ....(((((((((....)))---------(------(((((.((----((.((.(((.(.....))))))))))))))))...))))))................. ( -20.50, z-score = -0.49, R) >droYak2.chr2R 17967650 96 + 21139217 UCAAAGAUCCUUGGUAACAGAAGAGAUGGG------UUUUCAAA----GAACGCGUUUGAUGAUCAACCGUUUCGAAAGCAACGGACUUGCAAUUUAUCAAACCAA ...........((((........(((((.(------(((.....----)))).)))))(((((.((((((((.(....).)))))..)))....)))))..)))). ( -21.60, z-score = -0.97, R) >droSec1.super_9 1526676 96 + 3197100 UCAAAGGUCCUUGGCAACAGCAGAGGUGGG------UUUUCAGA----GAACGCGUUUGAUUAGCAACCGUUUCGAAAGCAACGGCCUUGCAAUUUAUCAAACCUC ........((((.((....)).)))).(.(------(((.....----)))).)(((((((..(((((((((.(....).)))))..)))).....)))))))... ( -25.80, z-score = -1.10, R) >droSim1.chr2R 16815706 96 + 19596830 UCAAAGGUCCUUGGCAACAGCAGAGGUGGG------UUUUCAGA----GAACGCGUUUGAUUAGCAACCGUUUCGAAAGCAACGGCCUUGCAAUUUAUCAAACCUC ........((((.((....)).)))).(.(------(((.....----)))).)(((((((..(((((((((.(....).)))))..)))).....)))))))... ( -25.80, z-score = -1.10, R) >droPer1.super_4 4136487 106 + 7162766 UCCCAGGUGGGGGACAACAGCAGAAACAGGGCAACAUUCCCGGACGGAGACCGAGCUCGAUUAGCAGCCGUUUCGAGAGCAACGGACUUCCCACUGAUUGAGCCUC ((((......))))..............(((.......)))...(((...))).((((((((((.(((((((.(....).))))).)).....))))))))))... ( -30.10, z-score = 0.24, R) >dp4.chr3 4864066 106 - 19779522 UCCCAGGUGGGGAACAACAGCAGAAACAGGGCAACACUCCCGGACGGAGACCGAGCUCGAUUAGCAGCCGUUUCGAGAGCAACGGACUUCCCACUGAUUGAGCCUC ((((.....))))................(....).((((.....)))).....((((((((((.(((((((.(....).))))).)).....))))))))))... ( -31.20, z-score = -0.21, R) >droGri2.scaffold_15245 13245973 83 + 18325388 ----------CCGAUGGCAAAAGGCAGAAG--------UUGAGAUC--GUGUGCGAACAAUUAGCAACCGUUUCGGAAGCAACGGAUUUGAAACUGAUUGCGU--- ----------......((((.((.((((.(--------((((..((--(....)))....)))))..(((((.(....).))))).))))...))..))))..--- ( -21.50, z-score = -0.82, R) >consensus UCAAAGGUCCUUGGCAACAGCAGAGAUGGG______UUUUCAGA____GAACGCGUUUGAUUAGCAACCGUUUCGAAAGCAACGGACUUGCAAUUUAUCGAACCUC ......................................................((((((((((((((((((.(....).)))))..)))...))))))))))... (-16.10 = -15.30 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:54 2011