
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,196,253 – 18,196,344 |
| Length | 91 |
| Max. P | 0.997550 |

| Location | 18,196,253 – 18,196,344 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.34 |
| Shannon entropy | 0.44928 |
| G+C content | 0.47103 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -11.31 |
| Energy contribution | -11.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18196253 91 + 21146708 AAGAGCCAACAACCUGUUGC-----------CUGAUUGUUA-UGCGACGGACGUGACCGAAAAUACUGUCACGUAAUCCGCAAACUCCCACAGAAAUGUUGGU------- ....(((((((..(((((((-----------.(((...)))-.)))))))(((((((.(......).)))))))......................)))))))------- ( -26.50, z-score = -2.96, R) >droSim1.chr2R 16793057 91 + 19596830 AAGAGCCAACAACCUGUUGC-----------CUGAUUGUUA-UGCGACGGACGUGACCGAAAAUACUGUCACGUAAUCCGCAAACUCCCACAGAAUUGUUGGU------- ....((((((((.(((((((-----------.(((...)))-.)))))))(((((((.(......).))))))).....................))))))))------- ( -27.10, z-score = -3.05, R) >droSec1.super_9 1505863 91 + 3197100 AAGAGCCAACAACCUGUUGC-----------CUGAUUGUUA-UGCGACGGACGUGACCGAAAAUACUGUCACGUAAUCCGCAAACUCCCACAGAAAUGUUGGU------- ....(((((((..(((((((-----------.(((...)))-.)))))))(((((((.(......).)))))))......................)))))))------- ( -26.50, z-score = -2.96, R) >droYak2.chr2R 17947214 100 + 21139217 AAGAGCCAACAACCUGUUGCC--------AGUUGAUUGUUA-UGCGAUGGACGUGACCGAAAAUACUGUCACGUAAUCCGCUAACUCCCACAGAAAUGUUGUGUGUGCU- ...(((((((((((((....)--------))......((((-.(((....(((((((.(......).)))))))....)))))))............))))).)).)))- ( -24.80, z-score = -1.44, R) >droEre2.scaffold_4845 12336409 90 + 22589142 AAGAGCCAACAACCUGUUGC-----------CUGAUUGUUA-UGCGACGGACGUGACCGAAAAUACUGUCACGUAAUCCGCAAACUCCCACAGAA-UGUGGCC------- ....((((.((..((((.(.-----------..((..(((.-((((....(((((((.(......).)))))))....)))))))))).))))..-)))))).------- ( -24.70, z-score = -2.43, R) >droAna3.scaffold_13266 2210588 98 + 19884421 AAGAGCCAACAACCUGUUGCCAC----UCGCCUGAUUGCUA-UGCAAGGGACGUGACCGAAAAUACUGUCACGUAAUCGGCCAACUCCCACAGAAGUGUGGCU------- ..................(((((----.(..(((.((((..-.))))((((..((.((((...(((......))).)))).))..)))).)))..).))))).------- ( -28.20, z-score = -1.41, R) >dp4.chr3 4840114 109 - 19779522 AAGAGCCAACAACCUGUGGCUUUAGCUUUAGCUGAUUGCUA-UGCAUCGGACGUGACCGAAAAUAUUGUCACGUAAUCCGUGGACUCCCACUGAAGUGUGGCUGUCGAUU .(((((((........)))))))(((....)))(((.((((-(((.(((((((((((..........))))))...)))((((....)))).)).))))))).))).... ( -37.50, z-score = -3.41, R) >droPer1.super_4 4112304 102 + 7162766 AAGAGCCAACAACCUGUGGCUUUAGCUUUAGCUGAUUGCUA-UGCAUCGGACGUGACCGAAAAUAUUGUCACGUAAUCCGUGGACUCCCACAGAAGUGUGGCU------- .(((((((........)))))))(((....)))....((((-(((.(((((((((((..........))))))...)))((((....)))).)).))))))).------- ( -35.00, z-score = -3.15, R) >droWil1.scaffold_180697 3962173 99 - 4168966 AUGAGUUUUCAACCUGUUGUUGG----UUUUUUGAUUUCCAAUGCAUUGAACGUGCCAAUAAAUAUUGUCACGUCCCACACACACACACACUCAGAGAUUAGU------- .(((((..((((..(((((..((----((....))))..)))))..))))(((((.((((....)))).)))))...............))))).........------- ( -18.30, z-score = -0.15, R) >consensus AAGAGCCAACAACCUGUUGC___________CUGAUUGUUA_UGCGACGGACGUGACCGAAAAUACUGUCACGUAAUCCGCAAACUCCCACAGAAAUGUUGCU_______ .............((((...................(((....))).((((((((((.(......).)))))))...))).........))))................. (-11.31 = -11.16 + -0.16)



| Location | 18,196,253 – 18,196,344 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.34 |
| Shannon entropy | 0.44928 |
| G+C content | 0.47103 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.74 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18196253 91 - 21146708 -------ACCAACAUUUCUGUGGGAGUUUGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCGUCGCA-UAACAAUCAG-----------GCAACAGGUUGUUGGCUCUU -------.(((.((....)))))((((.(((((...(((((((.(......).)))))))...)))))-((((((((.(-----------....).)))))))))))).. ( -31.40, z-score = -3.33, R) >droSim1.chr2R 16793057 91 - 19596830 -------ACCAACAAUUCUGUGGGAGUUUGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCGUCGCA-UAACAAUCAG-----------GCAACAGGUUGUUGGCUCUU -------.(((.((....)))))((((.(((((...(((((((.(......).)))))))...)))))-((((((((.(-----------....).)))))))))))).. ( -31.60, z-score = -3.36, R) >droSec1.super_9 1505863 91 - 3197100 -------ACCAACAUUUCUGUGGGAGUUUGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCGUCGCA-UAACAAUCAG-----------GCAACAGGUUGUUGGCUCUU -------.(((.((....)))))((((.(((((...(((((((.(......).)))))))...)))))-((((((((.(-----------....).)))))))))))).. ( -31.40, z-score = -3.33, R) >droYak2.chr2R 17947214 100 - 21139217 -AGCACACACAACAUUUCUGUGGGAGUUAGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCAUCGCA-UAACAAUCAACU--------GGCAACAGGUUGUUGGCUCUU -(((.((.(((((.......(((..((((((((...(((((((.(......).)))))))...)))).-))))..))).((--------(....)))))))))))))... ( -32.70, z-score = -3.65, R) >droEre2.scaffold_4845 12336409 90 - 22589142 -------GGCCACA-UUCUGUGGGAGUUUGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCGUCGCA-UAACAAUCAG-----------GCAACAGGUUGUUGGCUCUU -------..((((.-....))))((((.(((((...(((((((.(......).)))))))...)))))-((((((((.(-----------....).)))))))))))).. ( -34.30, z-score = -3.89, R) >droAna3.scaffold_13266 2210588 98 - 19884421 -------AGCCACACUUCUGUGGGAGUUGGCCGAUUACGUGACAGUAUUUUCGGUCACGUCCCUUGCA-UAGCAAUCAGGCGA----GUGGCAACAGGUUGUUGGCUCUU -------(((((((..(((((((((..(((((((.(((......)))...)))))))..)))).(((.-..((......))..----...)))))))).)).)))))... ( -35.90, z-score = -2.21, R) >dp4.chr3 4840114 109 + 19779522 AAUCGACAGCCACACUUCAGUGGGAGUCCACGGAUUACGUGACAAUAUUUUCGGUCACGUCCGAUGCA-UAGCAAUCAGCUAAAGCUAAAGCCACAGGUUGUUGGCUCUU ..(((((((((.(((....)))((......((((...((((((..........))))))))))..((.-((((.....))))..)).....))...)))))))))..... ( -32.70, z-score = -2.28, R) >droPer1.super_4 4112304 102 - 7162766 -------AGCCACACUUCUGUGGGAGUCCACGGAUUACGUGACAAUAUUUUCGGUCACGUCCGAUGCA-UAGCAAUCAGCUAAAGCUAAAGCCACAGGUUGUUGGCUCUU -------(((((((..(((((((.......((((...((((((..........))))))))))..((.-((((.....))))..)).....))))))).)).)))))... ( -34.70, z-score = -3.04, R) >droWil1.scaffold_180697 3962173 99 + 4168966 -------ACUAAUCUCUGAGUGUGUGUGUGUGUGGGACGUGACAAUAUUUAUUGGCACGUUCAAUGCAUUGGAAAUCAAAAAA----CCAACAACAGGUUGAAAACUCAU -------.........(((((......((((((.(((((((.((((....)))).))))))).))))))............((----((.......))))....))))). ( -23.50, z-score = -0.68, R) >consensus _______ACCAACAUUUCUGUGGGAGUUUGCGGAUUACGUGACAGUAUUUUCGGUCACGUCCGUCGCA_UAACAAUCAG___________GCAACAGGUUGUUGGCUCUU .....................((((((..((((...(((((((..........)))))))...))))..((((((((...................)))))))))))))) (-19.64 = -19.74 + 0.10)

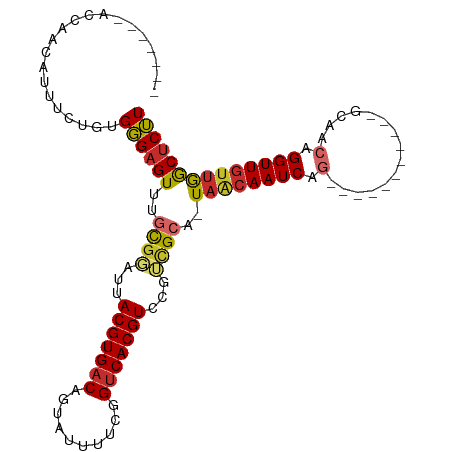
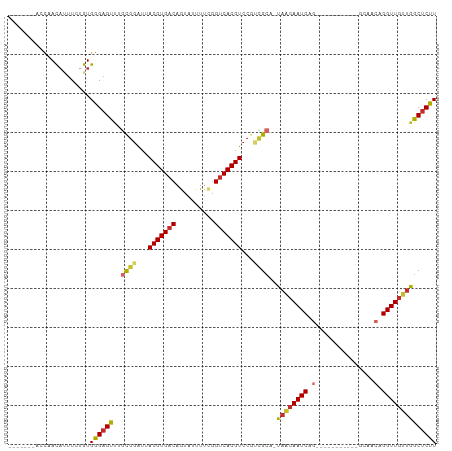
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:52 2011