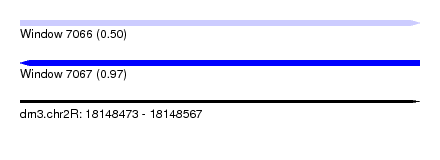
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,148,473 – 18,148,567 |
| Length | 94 |
| Max. P | 0.968640 |

| Location | 18,148,473 – 18,148,567 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.31660 |
| G+C content | 0.42931 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
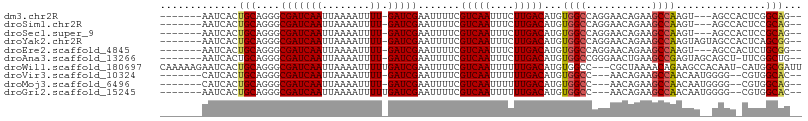
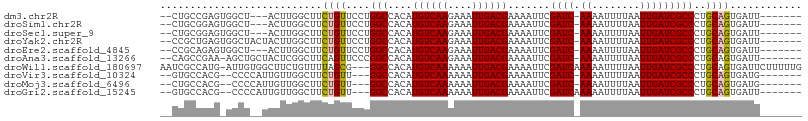
>dm3.chr2R 18148473 94 + 21146708 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGU---AGCCACUCGGCAG-- -------.....((((..(((((((((........))-))))........(((((....))))).....)))..)..)))..(((.(((---....))).)))..-- ( -24.70, z-score = -1.69, R) >droSim1.chr2R 16748047 94 + 19596830 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGU---AGCCACUCCGCAG-- -------.....((((.((((((((((........))-))))).......(((((....)))))..(((((..((........))....---.))))))))))))-- ( -26.50, z-score = -2.48, R) >droSec1.super_9 1458765 94 + 3197100 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGU---AGCCACUCCGCAG-- -------.....((((.((((((((((........))-))))).......(((((....)))))..(((((..((........))....---.))))))))))))-- ( -26.50, z-score = -2.48, R) >droYak2.chr2R 17898000 97 + 21139217 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGUAGUAGCCACUCAGCGG-- -------....(((((..(((((((((........))-)))).....((((((((....))))).(((........)))))))))..))))).((......))..-- ( -25.40, z-score = -1.75, R) >droEre2.scaffold_4845 12289446 94 + 22589142 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGU---AGCCACUCUGCGG-- -------.....(((((((.(((((((........))-))))).......(((((....)))))..(((((..((........))....---.))))))))))))-- ( -28.40, z-score = -2.66, R) >droAna3.scaffold_13266 2157437 96 + 19884421 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCGGGAACUGAAGCCGAGUAGCAGCU-UUCGGCUG-- -------..(((...(..(((((((((........))-))))........(((((....))))).....)))..)...)))(((((((........-))))))).-- ( -28.20, z-score = -1.60, R) >droWil1.scaffold_180697 3888904 103 - 4168966 CAAAAAGAAUCACUGCAGGGCGAUCAAUUAAAAUUUUUGAUCGAAUUUUCGUCAAUUUUUUGACAUGUGGCC---CGCUAAAACAGAAGCCACAAU-CAUGGCGAUU .......((((...((.((((((((((.........))))))........(((((....))))).....)))---)))..........((((....-..)))))))) ( -28.10, z-score = -2.75, R) >droVir3.scaffold_10324 627039 92 + 1288806 -------CAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUUUUGACAUGUGGCC---AACAGAAGCCAACAAUGGGG--CGUGGCAC-- -------...(((.......(((((((........))-))))).......(((((....)))))..)))(((---(......(((........))--).))))..-- ( -25.30, z-score = -1.85, R) >droMoj3.scaffold_6496 2464027 92 + 26866924 -------CAUCACUGCAGGGCGAUCAAUUAAAAUUUU-GAUCGAAUUUUCGUCAAUUUUUUGACAUGUGGCC---AACAGAAGCCAACAAUGGGG--CGUGGCAG-- -------.....((((..(((((((((........))-))))........(((((....))))).....)))---.......(((........))--)...))))-- ( -26.60, z-score = -2.25, R) >droGri2.scaffold_15245 8349231 93 + 18325388 -------AAUCACUGCAGGGCGAUCAAUUAAAAUUUUUGAUCGAAUUUUCGUCAAUUUUUUGACAUGUGGCC---AACAGAAGCCAACAAUGGGG--CGUGGCAC-- -------...(((.......(((((((.........))))))).......(((((....)))))..)))(((---(......(((........))--).))))..-- ( -24.50, z-score = -1.56, R) >consensus _______AAUCACUGCAGGGCGAUCAAUUAAAAUUUU_GAUCGAAUUUUCGUCAAUUUCUUGACAUGUGGCCAGGAACAGAAGCCAAGUA__AGCC_CUCGGCAG__ .............(((....(((((((........)).))))).......(((((....)))))...((((...........))))...............)))... (-13.72 = -13.68 + -0.04)
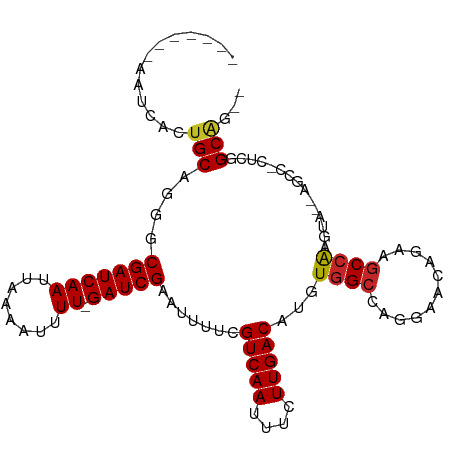
| Location | 18,148,473 – 18,148,567 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.31660 |
| G+C content | 0.42931 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.59 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
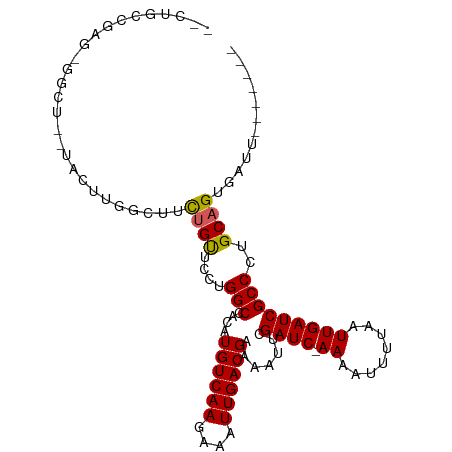
>dm3.chr2R 18148473 94 - 21146708 --CUGCCGAGUGGCU---ACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --..(((((((....---)))))))..((((....(((....((((((....)))))).......((((-((........)))))))))..)))).....------- ( -29.80, z-score = -3.55, R) >droSim1.chr2R 16748047 94 - 19596830 --CUGCGGAGUGGCU---ACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --((((((.((((((---(...(((....)))..))))))).((((((....))))))......(((((-((........)))))))..)))))).....------- ( -33.40, z-score = -4.30, R) >droSec1.super_9 1458765 94 - 3197100 --CUGCGGAGUGGCU---ACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --((((((.((((((---(...(((....)))..))))))).((((((....))))))......(((((-((........)))))))..)))))).....------- ( -33.40, z-score = -4.30, R) >droYak2.chr2R 17898000 97 - 21139217 --CCGCUGAGUGGCUACUACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --.(((((((.(((.......(((((.........)))))..((((((....)))))).......((((-((........))))))))))).)))))...------- ( -31.10, z-score = -4.02, R) >droEre2.scaffold_4845 12289446 94 - 22589142 --CCGCAGAGUGGCU---ACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --..((((.((((((---(...(((....)))..))))))).((((((....))))))......(((((-((........)))))))..)))).......------- ( -31.30, z-score = -3.78, R) >droAna3.scaffold_13266 2157437 96 - 19884421 --CAGCCGAA-AGCUGCUACUCGGCUUCAGUUCCCGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --..((((..-(((((((....)))...))))..))))(((.((((((....))))))......(((((-((........))))))).......)))...------- ( -27.00, z-score = -2.54, R) >droWil1.scaffold_180697 3888904 103 + 4168966 AAUCGCCAUG-AUUGUGGCUUCUGUUUUAGCG---GGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAAAAAUUUUAAUUGAUCGCCCUGCAGUGAUUCUUUUUG (((((((((.-...)))))..........(((---(((....((((((....)))))).......((((((.........)))))))))).))...))))....... ( -27.40, z-score = -2.37, R) >droVir3.scaffold_10324 627039 92 - 1288806 --GUGCCACG--CCCCAUUGUUGGCUUCUGUU---GGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUG------- --..((((.(--(......))))))..((((.---(((....((((((....)))))).......((((-((........)))))))))..)))).....------- ( -23.10, z-score = -1.85, R) >droMoj3.scaffold_6496 2464027 92 - 26866924 --CUGCCACG--CCCCAUUGUUGGCUUCUGUU---GGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUC-AAAAUUUUAAUUGAUCGCCCUGCAGUGAUG------- --..((((.(--(......))))))..((((.---(((....((((((....)))))).......((((-((........)))))))))..)))).....------- ( -23.10, z-score = -2.14, R) >droGri2.scaffold_15245 8349231 93 - 18325388 --GUGCCACG--CCCCAUUGUUGGCUUCUGUU---GGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAAAAAUUUUAAUUGAUCGCCCUGCAGUGAUU------- --..((((.(--(......))))))..((((.---(((....((((((....)))))).......((((((.........)))))))))..)))).....------- ( -22.30, z-score = -1.57, R) >consensus __CUGCCGAG_GGCU__UACUUGGCUUCUGUUCCUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUC_AAAAUUUUAAUUGAUCGCCCUGCAGUGAUU_______ ...........................((((....(((....((((((....)))))).......((((.((........)))))))))..))))............ (-14.57 = -14.59 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:47 2011