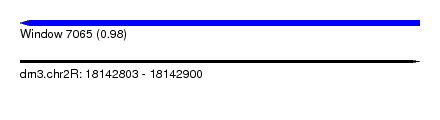
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,142,803 – 18,142,900 |
| Length | 97 |
| Max. P | 0.984873 |

| Location | 18,142,803 – 18,142,900 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Shannon entropy | 0.26626 |
| G+C content | 0.32846 |
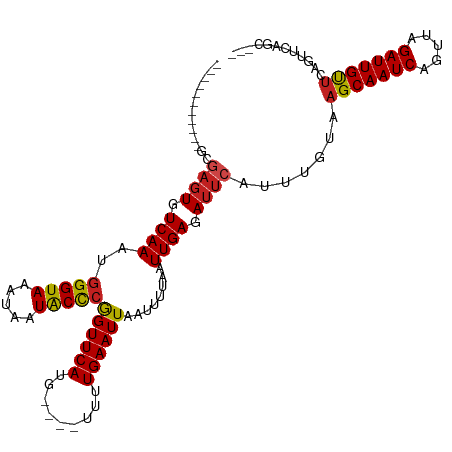
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984873 |
| Prediction | RNA |
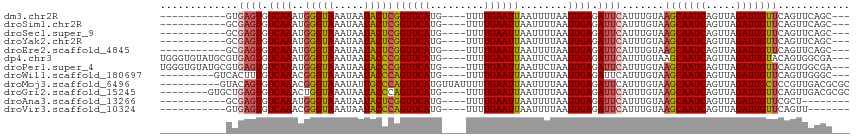
Download alignment: ClustalW | MAF
>dm3.chr2R 18142803 97 - 21146708 -----------GUGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC--- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))))).....(((((((.....))))))).........--- ( -21.50, z-score = -2.42, R) >droSim1.chr2R 16742444 97 - 19596830 -----------GCGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC--- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))).......(((((((.....))))))).......))--- ( -21.10, z-score = -2.15, R) >droSec1.super_9 1453183 97 - 3197100 -----------GCGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC--- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))).......(((((((.....))))))).......))--- ( -21.10, z-score = -2.15, R) >droYak2.chr2R 17892096 97 - 21139217 -----------GCGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC--- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))).......(((((((.....))))))).......))--- ( -21.10, z-score = -2.15, R) >droEre2.scaffold_4845 12283796 97 - 22589142 -----------GCGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC--- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))).......(((((((.....))))))).......))--- ( -21.10, z-score = -2.15, R) >dp4.chr3 4783454 108 + 19779522 UGGGUGUAUGCGUGAGUGUCAAAUGGGUAAAUAAUACCCGGUUCAUG----UUUUGAAUUAAUUCUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUACAGUGGCGA--- ........(((((((((.((((..(((((.....)))))((((((..----...))))))........)))).))))))((((..((((((.....))))))))))..))).--- ( -28.20, z-score = -2.23, R) >droPer1.super_4 4055035 108 - 7162766 UGGGUGUAUGCGUGAGUGUCAAAUGGGUAAAUAAUACCCGGUUCAUG----UUUUGAAUUAAUUCUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUGGCGA--- ........(((((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))))).....(((((((.....))))))).....))).--- ( -27.30, z-score = -2.06, R) >droWil1.scaffold_180697 284586 99 - 4168966 ---------GUCACUUUGUCAAACGGGUAAAUAAUACCCAGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUGGGC--- ---------(((....(.((((..(((((.....)))))((((((..----...))))))........)))).)..((.(((..(((((((.....)))))))))).)))))--- ( -20.10, z-score = -1.42, R) >droMoj3.scaffold_6496 2456602 105 - 26866924 ----------GUACAGUGUCAAACGGGUAAAUAUUGCCCAGUUCAUGUUAUUUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGCUCCGUUGACGCGC ----------.....(((((((..(((((.....)))))........((((...((((((............))))))...))))((((((.....))))))....))))))).. ( -26.60, z-score = -2.69, R) >droGri2.scaffold_15245 8340989 103 - 18325388 --------GUGCUGAGUGUCAAACUGGUAAAUAAUACCCAGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUGACGCGC --------(((((((((.((((...((((.....)))).((((((..----...))))))........)))).)))))......(((((((.....)))))))........)))) ( -23.80, z-score = -1.78, R) >droAna3.scaffold_13266 2151732 92 - 19884421 -----------GCGAGUGUCAAAUGGGUAAAUAAUACUCGGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCGCU-------- -----------..((((.((((..(((((.....)))))((((((..----...))))))........)))).))))....(..(((((((.....)))))))..).-------- ( -23.20, z-score = -3.01, R) >droVir3.scaffold_10324 620210 93 - 1288806 -----------GUGAGUGUCAAACGGGUAAAUAAUACCCAGUUCAUG----UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUU------- -----------((((((.((((..(((((.....)))))((((((..----...))))))........)))).)))))).....(((((((.....))))))).....------- ( -24.90, z-score = -4.29, R) >consensus ___________GCGAGUGUCAAAUGGGUAAAUAAUACCCGGUUCAUG____UUUUGAAUUAAUUUUAAUUGAGAUUCAUUUGUAAGCAAUCAGUUAGAUUGUUCAGUUCAGC___ .............((((.((((..(((((.....)))))((((((.........))))))........)))).)))).......(((((((.....)))))))............ (-19.88 = -19.76 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:45 2011