| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,126,670 – 18,126,776 |
| Length | 106 |
| Max. P | 0.691688 |

| Location | 18,126,670 – 18,126,776 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.46 |
| Shannon entropy | 0.72818 |
| G+C content | 0.47781 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -5.78 |
| Energy contribution | -5.34 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
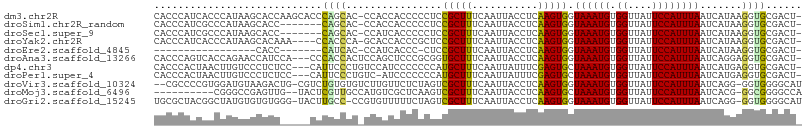
>dm3.chr2R 18126670 106 + 21146708 CACCCAUCACCCAUAAGCACCAAGCACCCAGCAC-CCACCACCCCCUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUAAGGUGCGACU- ................(((((..((.....))..-..(((((.....((((((...........)))))).....)))))..................)))))....- ( -23.00, z-score = -3.45, R) >droSim1.chr2R_random 2769587 99 + 2996586 CACCCAUCGCCCAUAAGCACC-------CAGCAC-CCACCACCCCCUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUAAGGUGCGACU- ......((((((....((...-------..))..-..(((((.....((((((...........)))))).....)))))..................)).))))..- ( -21.40, z-score = -2.93, R) >droSec1.super_9 1436955 99 + 3197100 CACCCAUCGCCCAUAAGCACC-------CAGCAC-CCAUCACCCCCUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUAAGGUGCGACU- ......((((......))...-------..((((-(.....................(((((......))))).((((((((.......)))))))).)))))))..- ( -20.70, z-score = -2.73, R) >droYak2.chr2R 17875193 102 + 21139217 CACCCAUCACCCAUAAGCACAAA----CCACCCA-GCACCACCCGCUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUAAGGUGCGACU- .......................----.......-(((((.................(((((......))))).((((((((.......)))))))).)))))....- ( -20.50, z-score = -2.36, R) >droEre2.scaffold_4845 12266872 81 + 22589142 -----------------CACC-------CAUCAC-CCAUCACCC-CUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUAAGGUGCGACU- -----------------....-------......-....((((.-............(((((......))))).((((((((.......)))))))).)))).....- ( -14.80, z-score = -1.69, R) >droAna3.scaffold_13266 2134857 104 + 19884421 CACCCAGUCACCAGAACCAUCCA---CCCACCACUCCAGCUCCCGCGGUGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAGGAGGUGCGACU- ......(.((((..((((((...---...((((((...((....))((((........))))...))))))....))))))..(((.........))))))))....- ( -24.20, z-score = -0.95, R) >dp4.chr3 4764685 104 - 19779522 CACCCACUAACUUGUCCCUCUCC---CAUUCCCUGUCCAUCCCCCCCAUGCUUUCAAUUAUUUCGAGUGCUAAAUGUGGUUAUUCCAUUUAAUCAUGAGGUGCGACU- ...........(((..((((...---.......................((.(((.........))).)).....(((((((.......)))))))))))..)))..- ( -14.90, z-score = -0.90, R) >droPer1.super_4 4036287 103 + 7162766 CACCCACUAACUUGUCCCUCUCC---CAUUCCCUGUC-AUCCCCCCCAUGCUUUCAAUUAUUUCGAGUGCUAAAUGUGGUUAUUCCAUUUAAUCAUGAGGUGCGACU- ...........(((..((((...---...........-...........((.(((.........))).)).....(((((((.......)))))))))))..)))..- ( -14.90, z-score = -0.76, R) >droVir3.scaffold_10324 596709 104 + 1288806 --CGCCCCGUGGAUGUAAGACUG-CGUCUGUGUGUCUUGUUCUCUAGUCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAGG-GGUGGGGCAU --.((((((..((((((....))-))))..)(((.((........)).)))........(((((...((((....((((.....))))...))))))-)))))))).. ( -32.40, z-score = -2.29, R) >droMoj3.scaffold_6496 2429618 95 + 26866924 ----------CGGGCCGAGUUG--UACUCGUUGCCAUGUCGCUCAAGUCGCUUUCAAUUACCUCAAGUGCUAAAUGUGGUUAUUCCAUUUAAUCACG-GGCGGGGCCA ----------..(((((((...--..))).........((((((.((.(((((...........)))))))....(((((((.......))))))))-))))))))). ( -29.30, z-score = -1.61, R) >droGri2.scaffold_15245 8318414 105 + 18325388 UGCGCUACGGCUAUGUGUGUGGG-UACUUGCC-CCGUGUUUUUCUAGUCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAGG-GGUGGGGCAU (((((....))............-..((..((-(((((..........)))...((.(((((......))))).))((((((.......))))))))-))..))))). ( -29.60, z-score = -0.45, R) >consensus CACCCAUCACCCAUAAGCACC_____CCCACCAC_CCAUCACCCCCUCCGCUUUCAAUUACCUCAAGUGGUAAAUGUGGUUAUUCCAUUUAAUCAUGAGGUGCGACU_ ............................((((................(((((...........))))).((((((.((....)))))))).......))))...... ( -5.78 = -5.34 + -0.44)
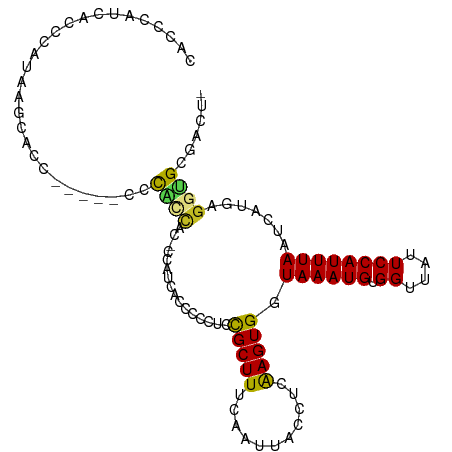

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:42 2011