| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,118,466 – 18,118,519 |
| Length | 53 |
| Max. P | 0.825659 |

| Location | 18,118,466 – 18,118,519 |
|---|---|
| Length | 53 |
| Sequences | 11 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Shannon entropy | 0.36382 |
| G+C content | 0.39350 |
| Mean single sequence MFE | -11.84 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.39 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
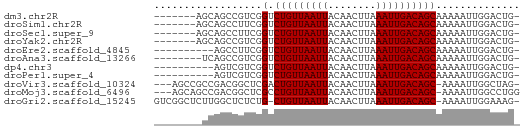
>dm3.chr2R 18118466 53 - 21146708 -------AGCAGCCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- -------..((((((...(.(((((((((........))))))))))......))).)))- ( -12.00, z-score = -1.82, R) >droSim1.chr2R 16718549 53 - 19596830 -------AGCAGCCUUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- -------..(((((....(.(((((((((........)))))))))).......)).)))- ( -11.10, z-score = -1.74, R) >droSec1.super_9 1428793 53 - 3197100 -------AGCAGCCUUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- -------..(((((....(.(((((((((........)))))))))).......)).)))- ( -11.10, z-score = -1.74, R) >droYak2.chr2R 17866678 53 - 21139217 -------AGCAGCCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- -------..((((((...(.(((((((((........))))))))))......))).)))- ( -12.00, z-score = -1.82, R) >droEre2.scaffold_4845 12257703 50 - 22589142 ----------AGCCUUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- ----------((((....(.(((((((((........)))))))))).......)).)).- ( -9.00, z-score = -1.45, R) >droAna3.scaffold_13266 2126282 52 - 19884421 --------UCAGCCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- --------.((((((...(.(((((((((........))))))))))......))).)))- ( -11.90, z-score = -2.37, R) >dp4.chr3 4755104 50 + 19779522 ----------AGUCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- ----------((((..(((((((((((((........))))))))).....)))))))).- ( -9.40, z-score = -1.38, R) >droPer1.super_4 4026653 50 - 7162766 ----------AGUCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG- ----------((((..(((((((((((((........))))))))).....)))))))).- ( -9.40, z-score = -1.38, R) >droVir3.scaffold_10324 582395 56 - 1288806 ---AGCCGCCGACGGCUCGACUGUUAAUUACAACUUAAAUUGACAGC-AAAAUUGGCUAG- ---(((((((...)))..(.(((((((((........))))))))))-......))))..- ( -16.70, z-score = -3.43, R) >droMoj3.scaffold_6496 2414054 57 - 26866924 ---AGCAGCCGACGGCUCGCCUGUUAAUUACAACUUAAAUUGACAGC-AAAAUUGGCCUGG ---..(((((((.((....))((((((((........))))))))..-....)))).))). ( -13.40, z-score = -0.90, R) >droGri2.scaffold_15245 8305526 58 - 18325388 GUCGGCUCUUGGCUCUCUG-CUGUUAAUUACAACUUAAAUUGACAGC-AAAAUUGGAAAG- (..(((.....)))..)((-(((((((((........))))))))))-)...........- ( -14.20, z-score = -2.74, R) >consensus ________GCAGCCGUCGGUCUGUUAAUUACAACUUAAAUUGACAGCAAAAAUUGGACUG_ ..................(.(((((((((........)))))))))).............. ( -7.39 = -7.39 + -0.00)

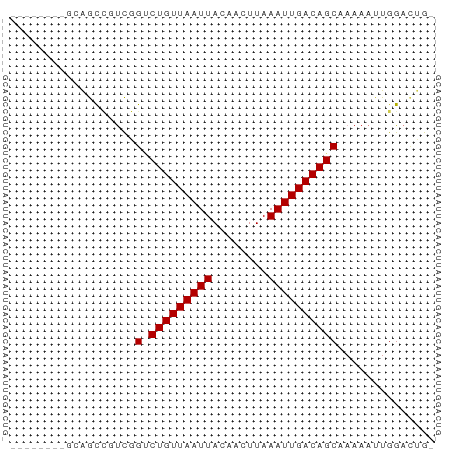
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:41 2011