
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,073,092 – 18,073,229 |
| Length | 137 |
| Max. P | 0.799611 |

| Location | 18,073,092 – 18,073,198 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.60 |
| Shannon entropy | 0.69776 |
| G+C content | 0.49167 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -13.72 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
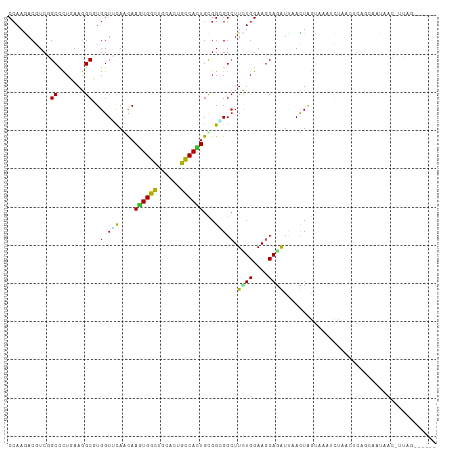
>dm3.chr2R 18073092 106 + 21146708 CCAAGACGUCGGCCCUCAAGGCGUGGUUUAGUAAGUGGCUGCACUGCCACUGUGGGGGCUUUUGGAAGCAGGUUCACUAGUAAAUCUAUGUGAGCAAUGACCUUAG------ (((((.....((((((((..((........)).((((((......)))))).)))))))))))))......(((((((((.....))).))))))...........------ ( -35.70, z-score = -0.86, R) >droGri2.scaffold_15112 3251396 110 + 5172618 CCAAAACAUCAGCGCUGAAGGCGUGGUGCAACAAGUGGUUGCAUUGCCACUGCGGCGGCUUCUGGAAGCAAAUUAAUUAGUCUAUUCUACUCUAGACGAAUAAGCAUUAC-- ...........((((((.((..(..(((((((.....)))))))..)..)).)))).((((....))))..........(((((........)))))......)).....-- ( -31.50, z-score = -1.18, R) >droMoj3.scaffold_6496 22302984 110 + 26866924 CCAAAACGUCUGCGUUGAAGGCGUGGUGCAACAAAUGGUUACAUUGCCACUGCGGUGGCUUCUGGAAGCAAAUCAAUUGAUAUUUUGUAAUCGUAACUUAAGAAUAUUAU-- .((..((((((.......))))))..))......(((((((((..(((((....)))))............(((....)))....)))))))))................-- ( -25.50, z-score = -0.38, R) >droVir3.scaffold_12875 17705796 108 - 20611582 CCAAAACGUCCGCACUGAAAGCCUGGUGCAACAAGUGGUUGCAUUGCCACUGUGGCGGCUUUUGGAAGCAAAUUAAUUAGUAU--UCAAAUAAGAAUUAAAGAGCUCUAG-- .......(..(((.(..((((((.((((((((.....))))))))(((.....)))))))))..)..))...((((((..(((--....)))..)))))).)..).....-- ( -30.60, z-score = -1.71, R) >droWil1.scaffold_180700 2296728 96 + 6630534 CCAAGACAUCCGCCCUGAAGGCCUGGUUUAACAAAUGGCUGCAUUGCCAUUGUGGAGGCUUCUGGAAGCAAAUUAAUUAAAUCCGUUCACACUUAC---------------- ....(((....(((.....)))..(((((((((((((((......)))))).))...((((....)))).......))))))).))).........---------------- ( -19.50, z-score = 0.86, R) >droPer1.super_37 482931 112 - 760358 CCAAGACUUCGGCUCUGAAGGCCUGGUUCAACAAGUGGCUGCAUUGCCAUUGCGGCGGCUUCUGGAAGCAGAUUAAUUAGCCACGGCCGCCCCUUUACAGUAUCAAUCUUAG ..((((.....(((.((((((............((((((......))))))(((((((((((((....))))......))))...))))).)))))).))).....)))).. ( -36.00, z-score = -0.90, R) >dp4.chr3 504470 112 - 19779522 CCAAGACUUCGGCUCUGAAGGCCUGGUUCAACAAGUGGCUGCAUUGCCAUUGCGGCGGCUUCUGGAAGCAGAUUAAUUAGCCACGGCCGCCCCUUUACAGUAUCAAUCUUAG ..((((.....(((.((((((............((((((......))))))(((((((((((((....))))......))))...))))).)))))).))).....)))).. ( -36.00, z-score = -0.90, R) >droAna3.scaffold_13266 9160578 96 - 19884421 CGAAAACAUCUGCCCUUAAAGCCUGGUUCAGUAAGUGGCUUCACUGUCACUGCGGUGGAUUCUGGAAGCAGAUGCACUAA-AAAUAUUAGGGAGUAG--------------- .........((((((((((...........(((.(((((......))))))))((((.((.(((....))))).))))..-.....)))))).))))--------------- ( -27.10, z-score = -1.17, R) >droEre2.scaffold_4845 12212507 99 + 22589142 CCAAGACGUCGGCCCUCAAGGCGUGGUUUAGCAAGUGGCUGCACUGUCACUGCGGGGGCUUCUGGAAGCAGGUCCACUAGUAAAUCUA-------AAUGACCUUAG------ (((.((....(((((((...((........)).((((((......))))))..))))))))))))....(((((...(((.....)))-------...)))))...------ ( -31.40, z-score = -0.22, R) >droYak2.chr2R 17819224 106 + 21139217 CCAAGACGUCGGCCCUCAAGGCGUGGUUCAGCAAGUGGCUGCACUGUCACUGCGGUGGCUUCUGGAAACAGGUUCACUAGUAAAUCUAAAUGAGCACUGUCCUUAG------ ....(((....(((.....)))(((.(((((((.(((((......))))))))(((((..((((....)))).)))))............))))))).))).....------ ( -33.80, z-score = -0.82, R) >droSec1.super_9 1383416 105 + 3197100 CCAAGACGUCGGCCCUCAAGGCGUGGUUUAGUAAGUGGCUGCACUGCCACUGUGGGGGCUUCUGGAAGCAGGUUCACUAGUAAAUCUAUGUGAGCAAUAAC-UUAG------ ..(((.....((((((((..((........)).((((((......)))))).)))))))).(((....)))(((((((((.....))).)))))).....)-))..------ ( -35.60, z-score = -1.15, R) >droSim1.chr2R 16676137 105 + 19596830 CCAAGACGUCGGCCCUCAAGGCGUGGUUUAGUAAGUGGCUGCACUGCCACUGUGGGGGCUUCUGGAAGCAGGUUCACUAGUAAAUCUGUGUGAGCAAUGAC-UUAG------ .......(((((((((((..((........)).((((((......)))))).)))))))).(((....)))(((((((((.....))).))))))...)))-....------ ( -38.00, z-score = -1.29, R) >consensus CCAAGACGUCGGCCCUGAAGGCGUGGUUCAACAAGUGGCUGCACUGCCACUGCGGCGGCUUCUGGAAGCAGAUUAACUAGUAAAUCUAACUCAGCAAUAAC_UUAG______ ...........((.......))..(.(((....((((((......))))))...))).).((((....))))........................................ (-13.72 = -12.98 + -0.75)

| Location | 18,073,124 – 18,073,229 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.49 |
| Shannon entropy | 0.70342 |
| G+C content | 0.38940 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -12.06 |
| Energy contribution | -11.34 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.799611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
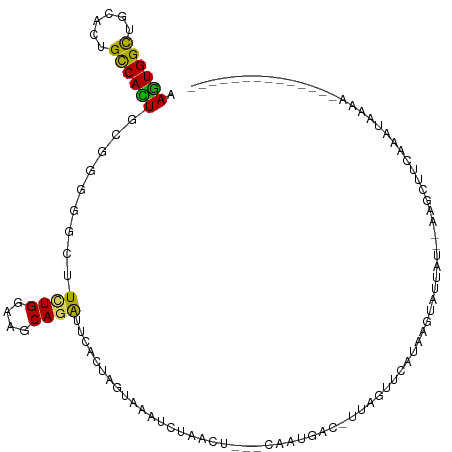
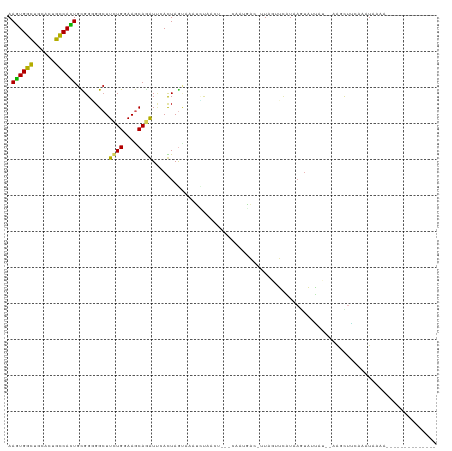
>dm3.chr2R 18073124 105 + 21146708 AAGUGGCUGCACUGCCACUGUGGGGGCUUUUGGAAGCAGGUUCACUAGUAAAUCUAUGUGAGCAAUGACCUUAGUUAUUAAGUAUGAUUUAAGCUUCAAAUAAAA-------------- .((((((......))))))...(((((((......((..(((((((((.....))).))))))((((((....))))))..)).......)))))))........-------------- ( -29.72, z-score = -1.45, R) >droGri2.scaffold_15112 3251428 95 + 5172618 AAGUGGUUGCAUUGCCACUGCGGCGGCUUCUGGAAGCAAAUUAAUUAGUCUAUUCUACU---CUAGACGAAUA-AGCAUUACUCUUU----AUUUGUAAGUAA---------------- .((((((......))))))((.((.((((....))))..........(((((.......---.))))).....-.)).((((.....----....))))))..---------------- ( -19.60, z-score = -0.01, R) >droWil1.scaffold_180700 2296760 94 + 6630534 AAAUGGCUGCAUUGCCAUUGUGGAGGCUUCUGGAAGCAAAUUAAUUAAAUCCGUUCACA---CUUACCAUCUACAGUAU---UCUUA---AGUUUAUAUGUAA---------------- .((((((......))))))(((((((....((..(((..(((.....)))..)))..))---....)).))))).....---.....---.............---------------- ( -16.40, z-score = 0.15, R) >droPer1.super_37 482963 109 - 760358 AAGUGGCUGCAUUGCCAUUGCGGCGGCUUCUGGAAGCAGAUUAAUUAGCCACGGCCGC--------CCCUUUACAGUAUCAAUCUUAG--AAUUUAAAUAUCCAUAGCUUUUAAUAAAA .((((((......))))))(((((((((((((....))))......))))...)))))--------..................((((--((.(((........))).))))))..... ( -25.40, z-score = -0.13, R) >dp4.chr3 504502 109 - 19779522 AAGUGGCUGCAUUGCCAUUGCGGCGGCUUCUGGAAGCAGAUUAAUUAGCCACGGCCGC--------CCCUUUACAGUAUCAAUCUUAG--AAUUUAAAUAUCCAUAGCUUUUAAUAAAA .((((((......))))))(((((((((((((....))))......))))...)))))--------..................((((--((.(((........))).))))))..... ( -25.40, z-score = -0.13, R) >droAna3.scaffold_13266 9160610 91 - 19884421 AAGUGGCUUCACUGUCACUGCGGUGGAUUCUGGAAGCAGAUGCACUAA-AAAUAUU-------AGGGAG--UAGUUUAUAAGCAUAA----AGUUUAAAAUAAAA-------------- .((((((......))))))..((((.((.(((....))))).))))..-.......-------......--..((((.(((((....----.)))))))))....-------------- ( -18.60, z-score = -0.84, R) >droEre2.scaffold_4845 12212539 98 + 22589142 AAGUGGCUGCACUGUCACUGCGGGGGCUUCUGGAAGCAGGUCCACUAGUAAAUCUA-------AAUGACCUUAGUUCUUAAGUAUAAUAUAAGCAUCAAAUAAAA-------------- .((((((......))))))((....((((..(((...(((((...(((.....)))-------...)))))...)))..)))).........))...........-------------- ( -20.52, z-score = 0.13, R) >droYak2.chr2R 17819256 105 + 21139217 AAGUGGCUGCACUGUCACUGCGGUGGCUUCUGGAAACAGGUUCACUAGUAAAUCUAAAUGAGCACUGUCCUUAGUUCUUAAGUAUAAUUUAAGCUUCAAAUAAAC-------------- .((((((......))))))(((((((..((((....)))).)))))............((((.((((....)))).))))............))...........-------------- ( -25.20, z-score = -1.22, R) >droSec1.super_9 1383448 104 + 3197100 AAGUGGCUGCACUGCCACUGUGGGGGCUUCUGGAAGCAGGUUCACUAGUAAAUCUAUGUGAGCAAUAAC-UUAGUUCUUAAGUAUGAUUUAAGCUUCAAAUAAAA-------------- .((((((......))))))...((((((((((....)))(((((((((.....))).))))))....((-((((...)))))).......)))))))........-------------- ( -30.60, z-score = -1.97, R) >droSim1.chr2R 16676169 104 + 19596830 AAGUGGCUGCACUGCCACUGUGGGGGCUUCUGGAAGCAGGUUCACUAGUAAAUCUGUGUGAGCAAUGAC-UUAGUUCUUAAGUAUGAUUUAAGCUUCAAAUAAAA-------------- .((((((......))))))...((((((((((....)))(((((((((.....))).))))))....((-((((...)))))).......)))))))........-------------- ( -30.40, z-score = -1.43, R) >consensus AAGUGGCUGCACUGCCACUGCGGGGGCUUCUGGAAGCAGAUUCACUAGUAAAUCUAACU___CAAUGAC_UUAGUUCAUAAGUAUUAU__AAGCUUCAAAUAAAA______________ .((((((......)))))).........((((....))))............................................................................... (-12.06 = -11.34 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:39 2011