| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,058,652 – 18,058,747 |
| Length | 95 |
| Max. P | 0.641399 |

| Location | 18,058,652 – 18,058,747 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Shannon entropy | 0.40817 |
| G+C content | 0.65771 |
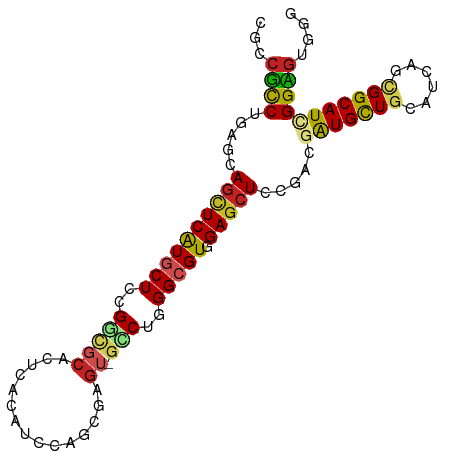
| Mean single sequence MFE | -45.64 |
| Consensus MFE | -21.31 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
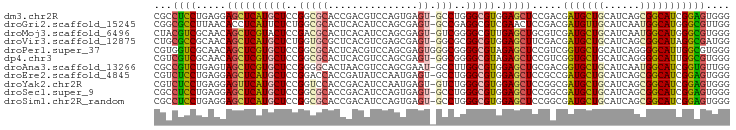
>dm3.chr2R 18058652 95 - 21146708 CGCCUCCUGAGGAGCUCAUGCUCCGGCGCACCGACGUCCAGUGAGU-GCCUGGGCGUGGAGCUCCGACGAUGCUGCAUCAGCGGCAUCGGAGUGGG (((.(((...(((((((((((.((((.((((..((.....))..))-)))))))))).)))))))...((((((((....)))))))))))))).. ( -53.00, z-score = -3.25, R) >droGri2.scaffold_15245 11823639 95 + 18325388 CGGCGCCUUAACACCUCAUUCUCUGGCGCACUCACAUCCAGCGAGU-GCCGAGGCGUCGAACUCCGACGAUGUUGCAUCAAUGGCAUGGGCGUUGG (((((((((....((.........)).((((((.(.....).))))-)).)))))))))....((((((....(((.......)))....)))))) ( -35.80, z-score = -1.67, R) >droMoj3.scaffold_6496 11732812 95 + 26866924 CUACGUCGCAACAGCUCGUACUCCGACGCACUCACAUCCAGCGAGU-GUCGGGGCGUUGAGCUGCGUCGAUGCUGCAUCAAUGGCAUGGGCGUGGG ((((.......(((((((..(((((((..((((.(.....).))))-)))))))...))))))).(((.(((((........))))).))))))). ( -40.50, z-score = -1.66, R) >droVir3.scaffold_12875 11722097 95 - 20611582 CUGCGCCGCAACAGCUCAUGCUCUGGUGCGCUCACGUCGAGCGAGU-GGCGCGGCGUGGAGCUUCGACGAUGCUGCAUCAGCGGCAUAGGCGAUGG (..((((((..((.(((..((......))((((.....))))))))-)..))))))..)..(.(((...(((((((....)))))))...))).). ( -44.30, z-score = -0.70, R) >droPer1.super_37 467696 96 + 760358 CGUGGUCGCAACAGCUCGUGCUCCGGCGCACUCACGUCCAGCGAGUGGGCGGGGCGUAGAGCUCCGUCGGUGCUGCAUCAGGGGCAUUGGCGUGGG .....((((...(((((..((((((.(.(((((.(.....).)))))).))))))...)))))..(((((((((.(....).))))))))))))). ( -45.90, z-score = -1.36, R) >dp4.chr3 489401 95 + 19779522 CGUCGUCGCAACAGCUCGUGCUCCGGCGCACUCACGUCCAGCGAGU-GGCGGGGCGUAGAGCUCCGUCGGUGCUGCAUCAGGGGCAUUGGCGUGGG .....((((...(((((..((((((.(.(((((.(.....).))))-))))))))...)))))..(((((((((.(....).))))))))))))). ( -43.40, z-score = -0.94, R) >droAna3.scaffold_13266 9145853 95 + 19884421 CGCCGUCUGAGUAGCUCGUGCUCCGGGGCACUAACGUCCAGCGAAU-GCCUUGGCGUGGAGCUGCGACGGUGCUGCAUCAAUGGCAUCGGUGUUGG .(((((.((((((((.((((((....)))))...((((((((..((-((....))))...)))).))))).))))).))))))))........... ( -43.80, z-score = -2.05, R) >droEre2.scaffold_4845 12198081 95 - 22589142 CGUCUCCUGAGGAGCUCAUGCUCCGGACCACCGAUAUCCAAUGAGU-GCCUGGGCGUGGAGCUCCGCCGAUGCUGCAUCAGCGGCAUCGGAGUGGG .....((...(((((((((((.((((..(((((........)).))-).)))))))).))))))).((((((((((....))))))))))...)). ( -45.70, z-score = -2.41, R) >droYak2.chr2R 17804683 95 - 21139217 CGUCUCCUGAGGAGUUCAUGCUCCGGUCCACCGACAUCCAAUGAGU-GUCUGGGCGUGGAGCUCCGGCGAUGCUGCAUCAGCGGCAUCGGAGUGGG ((.((((...(((((((((((.((((....))(((((.......))-))).)))))).)))))))...((((((((....)))))))))))))).. ( -45.00, z-score = -2.32, R) >droSec1.super_9 1368967 95 - 3197100 CGCCUCCUGAGGAGCUCAUGCUCCGGCGCACCGACAUCCAGUGAGU-GCCUGGGCGUGGAGCUCCGGCGAUGCUGCAUCAGCGGCAUCGGAGUGGG (((.(((...(((((((((((.((((.((((..((.....))..))-)))))))))).)))))))...((((((((....)))))))))))))).. ( -52.30, z-score = -2.86, R) >droSim1.chr2R_random 2750727 95 - 2996586 CGCCUCCUGAGGAGCUCAUGCUCCGGCGCACCGACAUCCAGUGAGU-GCCUGGGCGUGGAGCUCCGGCGAUGCUGCAUCAGCGGCAUCGGAGUGGG (((.(((...(((((((((((.((((.((((..((.....))..))-)))))))))).)))))))...((((((((....)))))))))))))).. ( -52.30, z-score = -2.86, R) >consensus CGCCGCCUGAGCAGCUCAUGCUCCGGCGCACUCACAUCCAGCGAGU_GCCUGGGCGUGGAGCUCCGACGAUGCUGCAUCAGCGGCAUCGGAGUGGG ...((((.....((((((((((..(((....................)))..))))).))))).....(((((((......))))))))))).... (-21.31 = -20.80 + -0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:37 2011