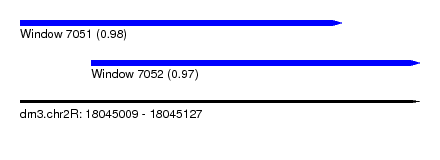
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 18,045,009 – 18,045,127 |
| Length | 118 |
| Max. P | 0.982227 |

| Location | 18,045,009 – 18,045,104 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.53 |
| Shannon entropy | 0.15237 |
| G+C content | 0.42342 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18045009 95 + 21146708 GAGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((((((--.....(((....)))..((((((...)))))).)))).....((....))(.((((((((((......)))))))))))))))....--- ( -28.10, z-score = -3.28, R) >droSim1.chr2R_random 2734006 95 + 2996586 GAGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((((((--.....(((....)))..((((((...)))))).)))).....((....))(.((((((((((......)))))))))))))))....--- ( -28.10, z-score = -3.28, R) >droSec1.super_9 1355425 95 + 3197100 GAGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((((((--.....(((....)))..((((((...)))))).)))).....((....))(.((((((((((......)))))))))))))))....--- ( -28.10, z-score = -3.28, R) >droYak2.chr2R 17790717 95 + 21139217 GUGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((....--.....))))(((.(((.....))))))....(((((......((....))(.((((((((((......))))))))))))))))...--- ( -26.80, z-score = -2.70, R) >droEre2.scaffold_4845 12184327 95 + 22589142 GUGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((....--.....))))(((.(((.....))))))....(((((......((....))(.((((((((((......))))))))))))))))...--- ( -26.80, z-score = -2.70, R) >droAna3.scaffold_13266 9131739 95 - 19884421 GAGCAACU--AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--- ((((((((--.....(((....)))..((((((...)))))).)))).....((....))(.((((((((((......)))))))))))))))....--- ( -28.10, z-score = -3.28, R) >droWil1.scaffold_180699 1422170 85 + 2593675 --------GAAAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUA--AAAAUUGAUGAAGCGCUUCU----- --------.......(((((((.(((.....))))))...............((....))...(((((((((.--...)))))))))))))....----- ( -22.90, z-score = -2.04, R) >droVir3.scaffold_12875 11704767 96 + 20611582 --GCAACUACAAAACGCGCGUGCUCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUA--AAAAUUGAUGAAGCGCUUCUGCUGC --.............((((..(((((.((((((...))))))((....)).......))))).(((((((((.--...)))))))))))))......... ( -22.80, z-score = -1.15, R) >consensus GAGCAACU__AAAACGCGCGUGCGCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU___ ...............(((....)))..((((((...))))))((((......((....))(.((((((((((......)))))))))))))))....... (-22.56 = -22.50 + -0.06)
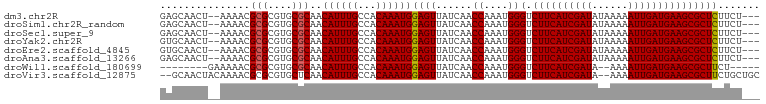
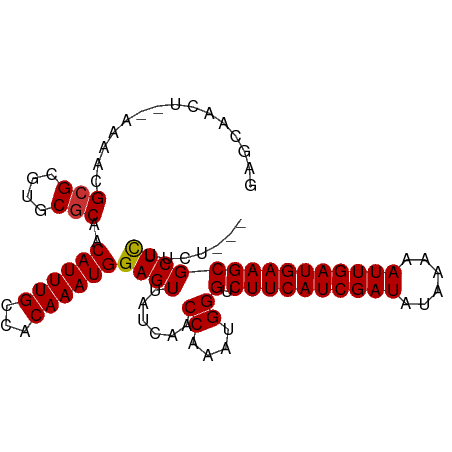
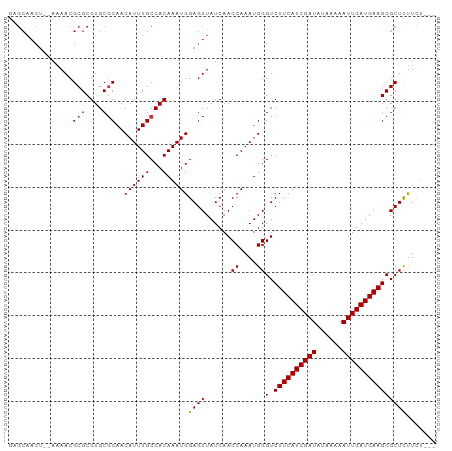
| Location | 18,045,030 – 18,045,127 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Shannon entropy | 0.33754 |
| G+C content | 0.41800 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 18045030 97 + 21146708 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCCUGUC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.40, z-score = -2.32, R) >droSim1.chr2R_random 2734027 97 + 2996586 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCCUGUC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.40, z-score = -2.32, R) >droSec1.super_9 1355446 97 + 3197100 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCCUGUC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.40, z-score = -2.32, R) >droYak2.chr2R 17790738 97 + 21139217 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCCUGUC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.40, z-score = -2.32, R) >droEre2.scaffold_4845 12184348 97 + 22589142 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCCUGUC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.40, z-score = -2.32, R) >droAna3.scaffold_13266 9131760 97 - 19884421 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU--CGGAGGCAAAACUUUGGCUUUGC--------- ((....((((((.(....(((((......((....))(.((((((((((......))))))))))))))))...--..).))))))......)).....--------- ( -26.80, z-score = -2.52, R) >droWil1.scaffold_180699 1422185 93 + 2593675 GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAU--AAAAAUUGAUGAAGCGCU--UCU--UCGGGGCAAAAAAGAAGCAUCUC--------- ((..(.((((((.(....(((((......((....))(.((((((((((--....))))))))))))))--)).--...)))))))...)..)).....--------- ( -25.80, z-score = -3.00, R) >droVir3.scaffold_12875 11704788 103 + 20611582 UCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAU--AAAAAUUGAUGAAGCGCUUCU---GCUGCGUAGAACUUGAGGCAACUUUGGGACGAC ....((((((...))))))..((..((........(.(.((((((((((--....))))))))))).)((((---((...))))))((.(((....))).))))..)) ( -25.40, z-score = -1.24, R) >droMoj3.scaffold_6496 11713861 106 - 26866924 UCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAU--AAAAAUUGAUGAAGCGCUUCUUCAGCUGCGUAGAACUCGAGGCAACUUUGGGAUGAC ....((((((...))))))..((((((..(((((...(.((((((((((--....)))))))))))((((((((.((...)).)))...)))))...))))))))))) ( -28.20, z-score = -1.83, R) >droGri2.scaffold_15245 11803773 99 - 18325388 UCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAU--AAAAAUUGAUGAAGCACU-------CUACGUAGAACUUGGGGCAACAUUGGGUUGUU ....((((((...)))))).......(((((.((((.(.((((((((((--....))))))))))).((-------(((.(.....).)))))...)))).))))).. ( -26.40, z-score = -2.23, R) >consensus GCAACAUUUGCCACAAAUGGAGUUAUCAACCAAAUGGGUCUUCAUCGAUAUAAAAAUUGAUGAAGCGCUCUUCU__CGGAGGCAAAACUUUGGCCUGUC_________ .........(((.....(((.........)))...((((((((((((((......)))))))))).)))).....................))).............. (-18.74 = -18.83 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:34 2011