
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,967,901 – 17,967,998 |
| Length | 97 |
| Max. P | 0.994229 |

| Location | 17,967,901 – 17,967,998 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.30144 |
| G+C content | 0.58400 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -29.06 |
| Energy contribution | -30.40 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
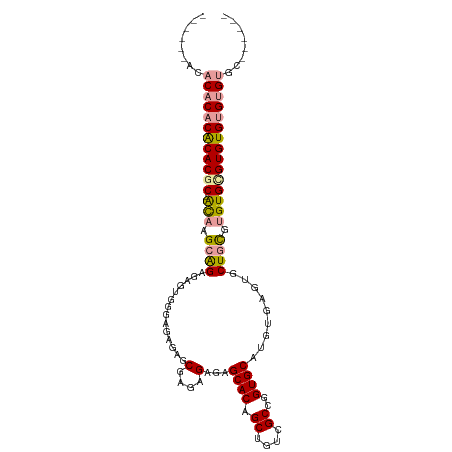
>dm3.chr2R 17967901 97 + 21146708 ------AAACACACGCACGC-GCCAGCAGUGAGUGGGAGAGAGCGAGAGAGAGCACAGCUGCCGCCGGUGCAUGUCAUUGCUGUGUGUGCGUGUGUGGGUGAAC---- ------..((((((((((((-((.((((((((((((.((...((........))....)).)))).........))))))))))))))))))))))........---- ( -45.00, z-score = -3.09, R) >droEre2.scaffold_4845 12106328 95 + 22589142 ------ACACACACACACGC-AUAAGCGGAGAGUGGGAGAGAGCGAGAGAGAGCACAGCUGUCGCCGGUGCAUGUGAGUGCUACGUGUGUGUGUGCGUGAAC------ ------.(((.(((((((((-((............((.((.(((.............))).)).)).((((((....)))).))))))))))))).)))...------ ( -35.62, z-score = -1.63, R) >droYak2.chr2R 9928514 108 - 21139217 ACACAAACACACACACACACGACAAGCAGAGAGUGUGAGAGAGCGAGAGAGAGCACAGCUGUCGCCGGUGCAUGUGAGUGCUGCCUGUGCGUGUGUGUGUGCGUGAAC ......((.(((((((((((((((.((((...(..((...(.((((.((.........)).)))))....))..).....)))).))).)))))))))))).)).... ( -43.40, z-score = -1.73, R) >consensus ______ACACACACACACGC_ACAAGCAGAGAGUGGGAGAGAGCGAGAGAGAGCACAGCUGUCGCCGGUGCAUGUGAGUGCUGCGUGUGCGUGUGUGUGUGC______ ......((((((((((((((....((((....(((.(...(.((((.((.........)).)))))..).))).....))))..)))))))))))))).......... (-29.06 = -30.40 + 1.34)


| Location | 17,967,901 – 17,967,998 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Shannon entropy | 0.30144 |
| G+C content | 0.58400 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -30.14 |
| Energy contribution | -29.92 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
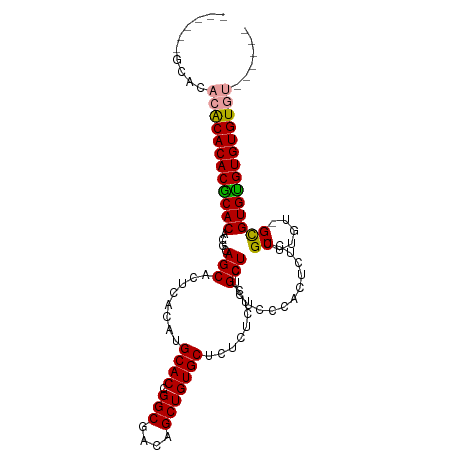
>dm3.chr2R 17967901 97 - 21146708 ----GUUCACCCACACACGCACACACAGCAAUGACAUGCACCGGCGGCAGCUGUGCUCUCUCUCGCUCUCUCCCACUCACUGCUGGC-GCGUGCGUGUGUUU------ ----........((((((((((.....(((......)))....(((.((((.(((......................))).)))).)-))))))))))))..------ ( -34.75, z-score = -2.18, R) >droEre2.scaffold_4845 12106328 95 - 22589142 ------GUUCACGCACACACACACGUAGCACUCACAUGCACCGGCGACAGCUGUGCUCUCUCUCGCUCUCUCCCACUCUCCGCUUAU-GCGUGUGUGUGUGU------ ------......(((((((((((((.(((........((((.(((....)))))))........)))).............((....-))))))))))))))------ ( -33.69, z-score = -3.04, R) >droYak2.chr2R 9928514 108 + 21139217 GUUCACGCACACACACACGCACAGGCAGCACUCACAUGCACCGGCGACAGCUGUGCUCUCUCUCGCUCUCUCACACUCUCUGCUUGUCGUGUGUGUGUGUGUUUGUGU ....(((((((((((((((.((((((((.........((((.(((....)))))))........(........).....)))))))))))))))))))))))...... ( -45.10, z-score = -3.12, R) >consensus ______GCACACACACACGCACACGCAGCACUCACAUGCACCGGCGACAGCUGUGCUCUCUCUCGCUCUCUCCCACUCUCUGCUUGU_GCGUGUGUGUGUGU______ ..........((((((((((((....(((........((((.(((....)))))))........)))..............((.....))))))))))))))...... (-30.14 = -29.92 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:29 2011