| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,932,098 – 17,932,206 |
| Length | 108 |
| Max. P | 0.967829 |

| Location | 17,932,098 – 17,932,206 |
|---|---|
| Length | 108 |
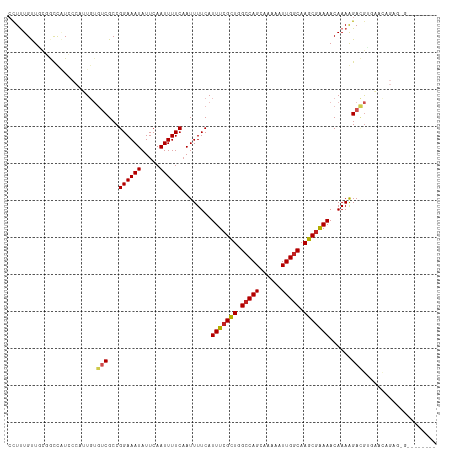
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Shannon entropy | 0.39325 |
| G+C content | 0.42653 |
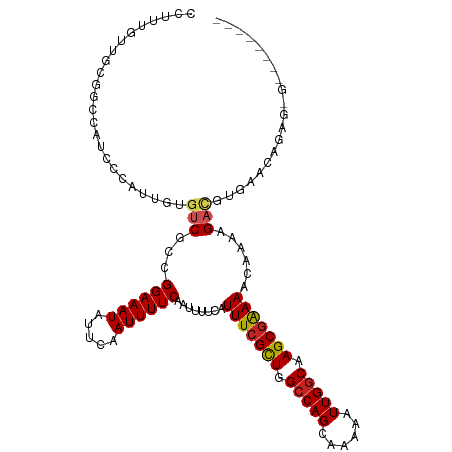
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -18.72 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17932098 108 + 21146708 CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAGGACGUGAACAGAGAG-------- (((((((((((((((......)).))))).((((((.....)))))).........((((((.(((((......))))).))))))))))))))..............-------- ( -34.20, z-score = -2.80, R) >droSim1.chr2R_random 2729367 108 + 2996586 CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAGGACGUGAACAGAGAG-------- (((((((((((((((......)).))))).((((((.....)))))).........((((((.(((((......))))).))))))))))))))..............-------- ( -34.20, z-score = -2.80, R) >droSec1.super_9 1244264 108 + 3197100 CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGAAAAAAUUGGCAAGCGAAAACAAAGGACGUGAACAGAGAG-------- (((((((((((((((......)).))))).((((((.....)))))).........((((((.(((((......))))).))))))))))))))..............-------- ( -33.90, z-score = -2.91, R) >droYak2.chr2R 9892136 104 - 21139217 CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGACAGCAAAAAUUGGCAAGCGAAAACAAAAGACGUGCACAG------------ ..(((((((((((((......)).))))).((((((.....)))))).........((((((...(((......)))...))))))))))))............------------ ( -22.00, z-score = 0.19, R) >droEre2.scaffold_4845 12067771 108 + 22589142 CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAAGACGUGAACAGAGAG-------- .((((((((((((((......)).))))).((((((.....))))))........(((((((.(((((......))))).)))))))............)))))))..-------- ( -32.40, z-score = -2.66, R) >droAna3.scaffold_13266 11302884 108 + 19884421 CUUUUGUUGCGGUCUUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCCAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAGGACGCUGCCAGGACG-------- ...........(((((..((..(((((...(((((........))))).......(((((((.(((((......))))).)))))))......)))))))..))))).-------- ( -32.20, z-score = -1.79, R) >dp4.chr3 15498810 116 + 19779522 CCUCUGCUCUUGCUCUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAAGACGUCAAUAGAGCGGGACAGAA ..((((.(((((((((...((((((((...((((((.....))))))........(((((((.(((((......))))).)))))))......)))).))))))))))))))))). ( -42.00, z-score = -4.87, R) >droPer1.super_4 3615061 116 - 7162766 CCUCUGCUCUUGCUCUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUUGCCAGCAAAAAUUGGCAAGCGAAAACAAAAGACGUCAAUAGAGCGGGACAAAA ....((.(((((((((...((((((((...((((((.....))))))........(((((((((((((......)))))))))))))......)))).)))))))))))))))... ( -41.60, z-score = -5.56, R) >droVir3.scaffold_12875 11560073 88 + 20611582 GCUUUGCUACCAUUGUG----U----CGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGUUGGCCAG---AAAUUGGCAAGCGAAAACAAAAGAAAAG----------------- ............((((.----.----....((((((.....))))))........(((((((.(((((---...))))).)))))))))))........----------------- ( -16.70, z-score = -0.24, R) >droMoj3.scaffold_6496 11542614 92 - 26866924 CCUUUGCUUGCAUUGUG----UGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAG---AAAUUGGCAAGCGAAAACAAAAGAUGAG----------------- ..........((((...----(((......((((((.....))))))........(((((((.(((((---...))))).))))))))))...))))..----------------- ( -22.20, z-score = -0.71, R) >droGri2.scaffold_15245 11662333 88 - 18325388 GCUUUGCUUCGAUUGUG----U----CGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAG---AAAUUGGCAAGCAGAAACAAAAGAAGAG----------------- ......((((..((((.----(----(...((((((.....))))))............(((.(((((---...))))).))).)).))))..))))..----------------- ( -21.10, z-score = -0.80, R) >consensus CCUUUGUUGCGGCCAUCCCAUUGUGUCGCCGGAAAUAUUCAAUUUUCAAUUUUCAUUUCGCUGGCCAGCAAAAAUUGGCAAGCGAAAACAAAAGACGUGAACAGAG_G________ ........................(((...((((((.....))))))........(((((((.(((((......))))).)))))))......))).................... (-18.72 = -19.04 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:24 2011