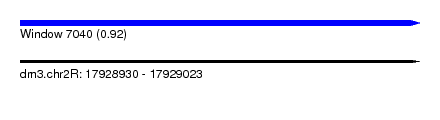
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,928,930 – 17,929,023 |
| Length | 93 |
| Max. P | 0.922882 |

| Location | 17,928,930 – 17,929,023 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.53480 |
| G+C content | 0.44656 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.89 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
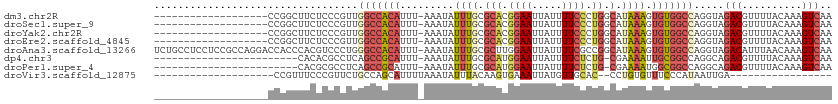
>dm3.chr2R 17928930 93 + 21146708 -------------------CCGGCUUCUCCCGUUGGCCACAUUU-AAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAA -------------------..(((((......((((((((((((-.......(((.((.((((.....)))).)))))..))))))))))))(((((...))))).))))).. ( -30.20, z-score = -2.98, R) >droSec1.super_9 1240815 93 + 3197100 -------------------CCGGCUUCUCCCGUUGGCCACAUUU-AAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAA -------------------..(((((......((((((((((((-.......(((.((.((((.....)))).)))))..))))))))))))(((((...))))).))))).. ( -30.20, z-score = -2.98, R) >droYak2.chr2R 9888768 93 - 21139217 -------------------CCGGCUUCUCCCGUUGGCCACAUUU-AAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAA -------------------..(((((......((((((((((((-.......(((.((.((((.....)))).)))))..))))))))))))(((((...))))).))))).. ( -30.20, z-score = -2.98, R) >droEre2.scaffold_4845 12064406 93 + 22589142 -------------------CCGGCUUCUCCCGUUGGCCACAUUU-AAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAA -------------------..(((((......((((((((((((-.......(((.((.((((.....)))).)))))..))))))))))))(((((...))))).))))).. ( -30.20, z-score = -2.98, R) >droAna3.scaffold_13266 11299377 112 + 19884421 UCUGCCUCCUCCGCCAGGACCACCCACGUCCCUGGGCCACAUUU-AAAUAUUUGCGCUUGGAAUUAUUUUCGCCGGCAUAAAGUGUGGCCAGGUAGACAUUUAACAAAGUCAA (((((((.......((((..(......)..))))((((((((((-.......(((...(((((....)))))...)))..)))))))))))))))))................ ( -32.70, z-score = -1.71, R) >dp4.chr3 15495320 87 + 19779522 ------------------------CACACGCCUCAGCCGCAUUU-AAAUAUUUGCGCAUGGAAUUAUUUUCUCUG-CGAAAAUUGCGGCCAGGCAGACGUUUUACAAAGUCAA ------------------------.....((((..((((((...-....((((.((((.((((.....)))).))-)).)))))))))).)))).(((..........))).. ( -27.91, z-score = -3.50, R) >droPer1.super_4 3611552 87 - 7162766 ------------------------CACGCGCCUCAGCCGCAUUU-AAAUAUUUGCGCAUGGAAUUAUUUUCUCUG-CGAAAAUGGCGGCCAGGCAGACGUUUUACAAAGUCAA ------------------------.....((((..(((((....-...(((((.((((.((((.....)))).))-)).)))))))))).)))).(((..........))).. ( -27.31, z-score = -2.29, R) >droVir3.scaffold_12875 11556972 74 + 20611582 --------------------CCGUUUCCCGUUCUGCCAGCAUUUUAAAUAUUUACAAGUGAAAUUAUGUUGCAC--CCUGUGUUUCCCAUAAUUGA----------------- --------------------...................((((((((....))).))))).(((((((..((((--...))))....)))))))..----------------- ( -6.30, z-score = 0.32, R) >consensus ___________________CCGGCUUCUCCCGUUGGCCACAUUU_AAAUAUUUGCGCACGGAAUUAUUUUCCCUGGCAUAAAGUGUGGCCAGGUAGACGUUUUACAAAGUCAA ..................................(((((((.........((((.(((.((((.....)))).)).).)))).))))))).....(((..........))).. (-10.01 = -10.89 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:24 2011