| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,629,220 – 3,629,324 |
| Length | 104 |
| Max. P | 0.999795 |

| Location | 3,629,220 – 3,629,320 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 62.63 |
| Shannon entropy | 0.60384 |
| G+C content | 0.48195 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -16.27 |
| Energy contribution | -19.15 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999795 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3629220 100 + 23011544 UUGAAAGAGAUUUUCUUGGCAUGAGCAC-ACUGCACUUAGCCCUCAUAAAGGGCCGAAUUUUCCUCAAAAUAAGUGUGGAAAGUUACGGCCCGCGGCCAAG ..............(((((((((((...-.(((....)))..)))))...((((((((((((((.((.......)).)))))))).))))))...)))))) ( -35.30, z-score = -3.46, R) >droSim1.chr2L 3588305 97 + 22036055 UCGAAAGAGAUUUUCUUGGCUUGAGCAC-ACUGCACUUAACCCUCAUGAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGUCGAG--- ((....))......((((((....((..-...))................(((((((((((((((((.......))))))))))).))))))))))))--- ( -39.30, z-score = -5.04, R) >droSec1.super_5 1730259 98 + 5866729 UCGAAAGAGAUUUUCUUGGCAUGAGCUCCACUGCACUUAAUCCUCACAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGUCGAG--- ((....))......((((((....((......))................(((((((((((((((((.......))))))))))).))))))))))))--- ( -40.20, z-score = -5.47, R) >droVir3.scaffold_12963 5085772 79 - 20206255 -----------CUACAACAAGUUUGCAA-AUUGAAAGGCGCCGCCAAAGGGGGCAGAGUU--CAAAAGGUUUGGCAUAGAGAGCGAGAGAGAG-------- -----------..........(((((..-.(((((....(((.((...)).)))....))--)))...(.....).......)))))......-------- ( -13.80, z-score = 0.60, R) >consensus UCGAAAGAGAUUUUCUUGGCAUGAGCAC_ACUGCACUUAACCCUCAUAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGUCGAG___ ........................((......))................(((((((((((((((((.......))))))))))).))))))......... (-16.27 = -19.15 + 2.88)

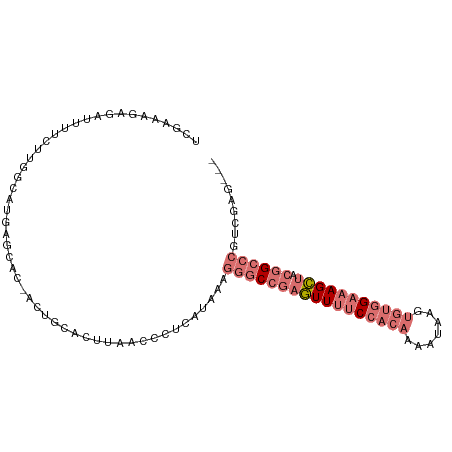

| Location | 3,629,220 – 3,629,320 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 62.63 |
| Shannon entropy | 0.60384 |
| G+C content | 0.48195 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -13.70 |
| Energy contribution | -15.08 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992855 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3629220 100 - 23011544 CUUGGCCGCGGGCCGUAACUUUCCACACUUAUUUUGAGGAAAAUUCGGCCCUUUAUGAGGGCUAAGUGCAGU-GUGCUCAUGCCAAGAAAAUCUCUUUCAA ((((((...((((((.((.(((((.((.......)).))))).))))))))...(((((.((..........-)).))))))))))).............. ( -34.70, z-score = -2.83, R) >droSim1.chr2L 3588305 97 - 22036055 ---CUCGACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUCAUGAGGGUUAAGUGCAGU-GUGCUCAAGCCAAGAAAAUCUCUUUCGA ---(((((.((((((.((.((((((((.......)))))))).)))))))).))..)))((((.((..(...-)..))..))))..((((.....)))).. ( -34.60, z-score = -3.21, R) >droSec1.super_5 1730259 98 - 5866729 ---CUCGACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUGUGAGGAUUAAGUGCAGUGGAGCUCAUGCCAAGAAAAUCUCUUUCGA ---((((..((((((.((.((((((((.......)))))))).))))))))....))))......(.(((...(....).))))..((((.....)))).. ( -31.30, z-score = -1.84, R) >droVir3.scaffold_12963 5085772 79 + 20206255 --------CUCUCUCUCGCUCUCUAUGCCAAACCUUUUG--AACUCUGCCCCCUUUGGCGGCGCCUUUCAAU-UUGCAAACUUGUUGUAG----------- --------.........((.......))........(((--((...((((((....)).))))...))))).-.(((((.....))))).----------- ( -11.60, z-score = -0.62, R) >consensus ___CUCGACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUAUGAGGGCUAAGUGCAGU_GUGCUCAUGCCAAGAAAAUCUCUUUCGA ..........(((...(((((((((((.......))))))))(((.(((((((...))))))).)))........)))...)))................. (-13.70 = -15.08 + 1.38)

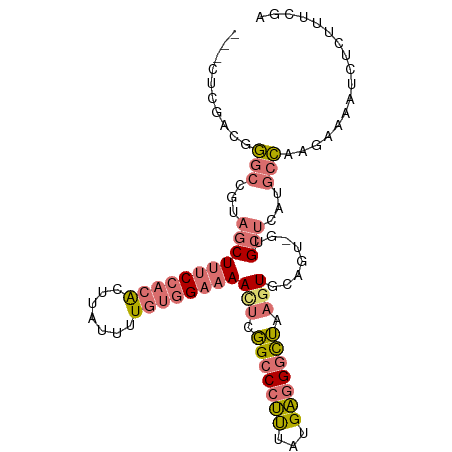

| Location | 3,629,224 – 3,629,324 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 64.44 |
| Shannon entropy | 0.56971 |
| G+C content | 0.48445 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -17.88 |
| Energy contribution | -20.00 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999652 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3629224 100 + 23011544 AAGAGAUUUUCUUGGCAUGAGCAC--ACUGCACUUAGCCCUCAUAAAGGGCCGAAUUUUCCUCAAAAUAAGUGUGGAAAGUUACGGCCCGCGGCCAAGCUUG ..........(((((((((((...--.(((....)))..)))))...((((((((((((((.((.......)).)))))))).))))))...)))))).... ( -35.70, z-score = -3.18, R) >droSim1.chr2L 3588309 97 + 22036055 AAGAGAUUUUCUUGGCUUGAGCAC--ACUGCACUUAACCCUCAUGAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGU---CGAGCUUG ..(((.....)))(((((((((..--...))................(((((((((((((((((.......))))))))))).)))))).)---)))))).. ( -40.00, z-score = -4.97, R) >droSec1.super_5 1730263 98 + 5866729 AAGAGAUUUUCUUGGCAUGAGCUCC-ACUGCACUUAAUCCUCACAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGU---CGAGCUUG ..........((((((....((...-...))................(((((((((((((((((.......))))))))))).))))))))---)))).... ( -38.30, z-score = -4.59, R) >droVir3.scaffold_12963 5085776 79 - 20206255 -------------AACAAGUUUGCAAAUUGAAAGGCGCCGCCAAAGGGGGCAGAGUU--CAAAAGGUUUGGCAUAGAGAGCGAGAGA--------GAGGUUG -------------..(((.((..(...(((((....(((.((...)).)))....))--)))....(((.((.......)).)))).--------.)).))) ( -15.50, z-score = 0.12, R) >consensus AAGAGAUUUUCUUGGCAUGAGCAC__ACUGCACUUAACCCUCAUAAAGGGCCGAGUUUUCCACAAAAUAAGUGUGGAAAGCUACGGCCCGU___CGAGCUUG .............(((....((.......))................(((((((((((((((((.......))))))))))).))))))........))).. (-17.88 = -20.00 + 2.12)


| Location | 3,629,224 – 3,629,324 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.44 |
| Shannon entropy | 0.56971 |
| G+C content | 0.48445 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -15.59 |
| Energy contribution | -17.78 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.995673 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3629224 100 - 23011544 CAAGCUUGGCCGCGGGCCGUAACUUUCCACACUUAUUUUGAGGAAAAUUCGGCCCUUUAUGAGGGCUAAGUGCAGU--GUGCUCAUGCCAAGAAAAUCUCUU ....((((((...((((((.((.(((((.((.......)).))))).))))))))...(((((.((..........--)).))))))))))).......... ( -34.90, z-score = -2.47, R) >droSim1.chr2L 3588309 97 - 22036055 CAAGCUCG---ACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUCAUGAGGGUUAAGUGCAGU--GUGCUCAAGCCAAGAAAAUCUCUU ....((((---(.((((((.((.((((((((.......)))))))).)))))))).))..)))((((.((..(...--)..))..))))((((.....)))) ( -33.90, z-score = -2.93, R) >droSec1.super_5 1730263 98 - 5866729 CAAGCUCG---ACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUGUGAGGAUUAAGUGCAGU-GGAGCUCAUGCCAAGAAAAUCUCUU ..(((((.---((((((((.((.((((((((.......)))))))).))))))))..(((...........)))))-.)))))......((((.....)))) ( -34.70, z-score = -2.85, R) >droVir3.scaffold_12963 5085776 79 + 20206255 CAACCUC--------UCUCUCGCUCUCUAUGCCAAACCUUUUG--AACUCUGCCCCCUUUGGCGGCGCCUUUCAAUUUGCAAACUUGUU------------- .......--------..............(((.(((....(((--((...((((((....)).))))...)))))))))))........------------- ( -10.60, z-score = -0.60, R) >consensus CAAGCUCG___ACGGGCCGUAGCUUUCCACACUUAUUUUGUGGAAAACUCGGCCCUUUAUGAGGGCUAAGUGCAGU__GUGCUCAUGCCAAGAAAAUCUCUU ....((((.....((((((.((.((((((((.......)))))))).))))))))....))))(((..(((((.....)))))...)))............. (-15.59 = -17.78 + 2.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:16 2011