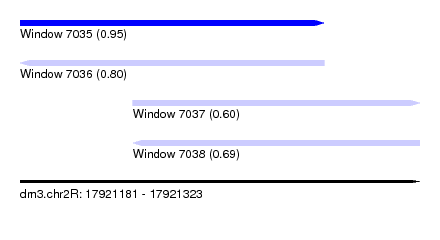
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,921,181 – 17,921,323 |
| Length | 142 |
| Max. P | 0.951773 |

| Location | 17,921,181 – 17,921,289 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.34084 |
| G+C content | 0.35239 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.44 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
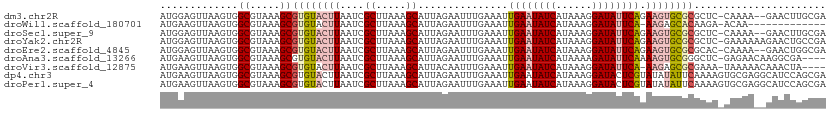
>dm3.chr2R 17921181 108 + 21146708 AUGGAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGAAGUGCGCGCUC-CAAAA--GAACUUGCGA ...(((((...(((.....(((((((((((....((.....))..............(((((((((......)))))))))))))))))))))-))...--.))))).... ( -32.60, z-score = -3.52, R) >droWil1.scaffold_180701 3308159 96 + 3904529 AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCA-AAGAGCACAAGA-ACAA------------- .....(((..(((((...(((((..........)))))...))..............(((((((((......))))))))-)....)))...)-))..------------- ( -20.40, z-score = -2.30, R) >droSec1.super_9 1232977 108 + 3197100 AUGGAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGAAGUGCGCGCUC-CAAAA--GAACUUGCGA ...(((((...(((.....(((((((((((....((.....))..............(((((((((......)))))))))))))))))))))-))...--.))))).... ( -32.60, z-score = -3.52, R) >droYak2.chr2R 9880731 110 - 21139217 AUGGAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGAAGUGCGCGCUC-GAAAAAAGAACUGCCGA .(((((((...(..((...(((((((((((....((.....))..............(((((((((......)))))))))))))))))))))-)..)....)))).))). ( -32.70, z-score = -3.49, R) >droEre2.scaffold_4845 12056460 108 + 22589142 AUGGAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGAAGUGCGCGCAC-CAAAA--GAACUGGCGA .(((.(((....))).....((((((((((....((.....))..............(((((((((......))))))))))))))))))).)-))...--.......... ( -29.10, z-score = -2.48, R) >droAna3.scaffold_13266 11291732 106 + 19884421 AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAAGAUAUUCAAAAGUGCGGGCUC-GAGAACAAGGCGA---- .....(((..((..((...(((.(((((((....((.....))..............(((((((((......)))))))))))))))).))))-)...))..)))..---- ( -25.90, z-score = -2.09, R) >droVir3.scaffold_12875 11545747 105 + 20611582 AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUACAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCA-AAGAGCGCGAAA-UAAAAACAAACUA---- .....(((((((((((.....))).))))))))(((((....((........))...(((((((((......))))))))-).))))).....-.............---- ( -21.20, z-score = -1.89, R) >dp4.chr3 15487059 111 + 19779522 AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUACUCGUAUAUAUUCAAAAGUGCGAGGCAUCCAGCGA .....(((((((((((.....))).))))))))((((..........((((((......)))).)).....((((.(((((((..........)))))))..)))))))). ( -25.40, z-score = -1.38, R) >droPer1.super_4 3603230 111 - 7162766 AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUACUCGUAUAUAUUCAAAAGUGCGAGGCAUCCAGCGA .....(((((((((((.....))).))))))))((((..........((((((......)))).)).....((((.(((((((..........)))))))..)))))))). ( -25.40, z-score = -1.38, R) >consensus AUGAAGUUAAGUGGCGUAAAGCGUGUACUUAAUCGCUUAAAGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGAAGUGCGCGCAC_CAAAA__GAACUAGCGA .(.(((((.....((...(((((..........)))))...)).....))))).)...((((((((......))))))))............................... (-16.64 = -16.44 + -0.20)
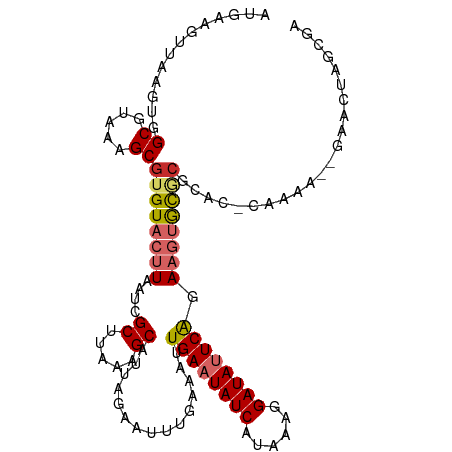
| Location | 17,921,181 – 17,921,289 |
|---|---|
| Length | 108 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Shannon entropy | 0.34084 |
| G+C content | 0.35239 |
| Mean single sequence MFE | -19.14 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.10 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17921181 108 - 21146708 UCGCAAGUUC--UUUUG-GAGCGCGCACUUCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUCCAU ....((((..--...((-(((((.(.((((.((((((((......))))))))..............(((.....)))...)))).).)))))))....))))........ ( -20.40, z-score = -1.79, R) >droWil1.scaffold_180701 3308159 96 - 3904529 -------------UUGU-UCUUGUGCUCUU-UGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU -------------..((-(...(((....(-((((((((......)))))))))..............((...(((((..........)))))...)))))..)))..... ( -15.90, z-score = -2.36, R) >droSec1.super_9 1232977 108 - 3197100 UCGCAAGUUC--UUUUG-GAGCGCGCACUUCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUCCAU ....((((..--...((-(((((.(.((((.((((((((......))))))))..............(((.....)))...)))).).)))))))....))))........ ( -20.40, z-score = -1.79, R) >droYak2.chr2R 9880731 110 + 21139217 UCGGCAGUUCUUUUUUC-GAGCGCGCACUUCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUCCAU ..(((.((((.......-))))(((.((((.((((((((......))))))))..............(((.....)))...))))...))).....)))............ ( -21.90, z-score = -2.39, R) >droEre2.scaffold_4845 12056460 108 - 22589142 UCGCCAGUUC--UUUUG-GUGCGCGCACUUCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUCCAU ..(((((...--..)))-))(((.(.((((.((((((((......))))))))..............(((.....)))...)))).).))).................... ( -19.80, z-score = -1.73, R) >droAna3.scaffold_13266 11291732 106 - 19884421 ----UCGCCUUGUUCUC-GAGCCCGCACUUUUGAAUAUCUUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU ----((((((((....)-))).........(((((((((......))))))))).....................))))((((((....((.....)).))))))...... ( -18.10, z-score = -2.17, R) >droVir3.scaffold_12875 11545747 105 - 20611582 ----UAGUUUGUUUUUA-UUUCGCGCUCUU-UGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUGUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU ----.(((.(((.((((-..((((.....(-((((((((......)))))))))..........(((....))).)))).)))).))).)))................... ( -17.00, z-score = -1.58, R) >dp4.chr3 15487059 111 - 19779522 UCGCUGGAUGCCUCGCACUUUUGAAUAUAUACGAGUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU (((((....((........((((((.......(((((((......)))))))...)))))).......))....)))))((((((....((.....)).))))))...... ( -19.36, z-score = -1.58, R) >droPer1.super_4 3603230 111 + 7162766 UCGCUGGAUGCCUCGCACUUUUGAAUAUAUACGAGUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU (((((....((........((((((.......(((((((......)))))))...)))))).......))....)))))((((((....((.....)).))))))...... ( -19.36, z-score = -1.58, R) >consensus UCGCUAGUUC__UUUUA_GAGCGCGCACUUCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCUUUAAGCGAUUAAGUACACGCUUUACGCCACUUAACUUCAU ................................(((((((......)))))))................((...(((((..........)))))...))............. (-12.27 = -12.10 + -0.17)
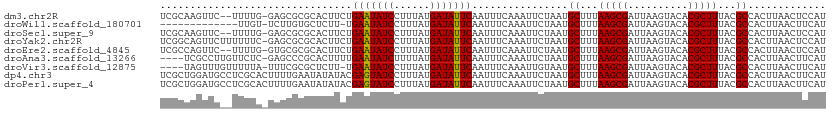
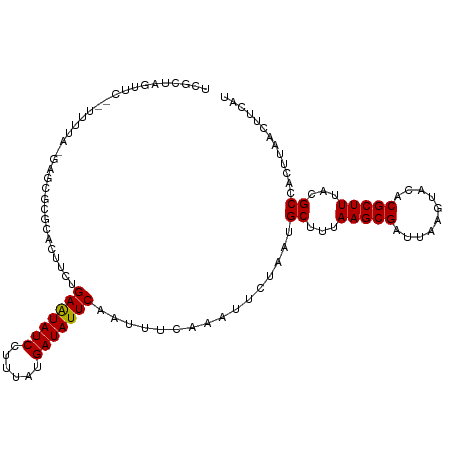
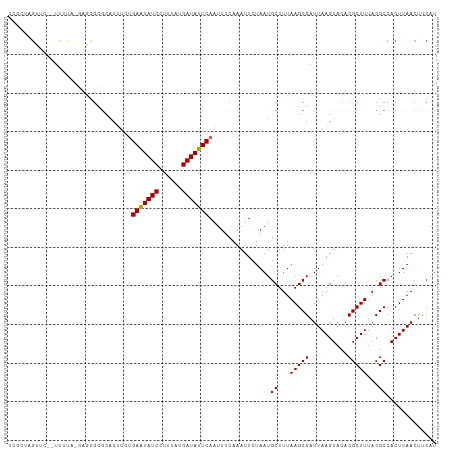
| Location | 17,921,221 – 17,921,323 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.42 |
| Shannon entropy | 0.61895 |
| G+C content | 0.35854 |
| Mean single sequence MFE | -16.17 |
| Consensus MFE | -7.30 |
| Energy contribution | -7.49 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17921221 102 + 21146708 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGA-AG---UGCGCGCUCCAAAA--GAACUUGCGAAAAA--AGAAUCCAUGCAAAACAAGGAGAACAGAGC--- .(((((...........(((((((((......))))))))).-))---)))..((((.....--...(((.......)--))..(((.((.....)).))).....))))--- ( -17.70, z-score = -0.98, R) >droWil1.scaffold_180701 3308199 90 + 3904529 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAAAGAG---CACAAGAACAAAAAAAAAACCCAAACUAGA--CUGAAACGUGAAAGA------------------ .................(((((((((......)))))))))....---..............................--...............------------------ ( -8.80, z-score = -1.10, R) >droSec1.super_9 1233017 102 + 3197100 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGA-AG---UGCGCGCUCCAAAA--GAACUUGCGAAAAA--CGAAUCCUUGCAAAACAAGGAGAGCAGAGC--- .(((((...........(((((((((......))))))))).-))---)))..((((.....--.......((.....--))..((((((.....)))))))))).....--- ( -22.60, z-score = -1.77, R) >droYak2.chr2R 9880771 104 - 21139217 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGA-AG---UGCGCGCUCGAAAAAAGAACUGCCGAGAAA--AGAAUCCAUGCAAAACAAGACAAGCGAAGC--- .((((............(((((((((......))))))))).-..---......((((.............))))...--.......))))...................--- ( -15.22, z-score = 0.30, R) >droEre2.scaffold_4845 12056500 105 + 22589142 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGA-AG---UGCGCGCACCAAAA--GAACUGGCGAGAAA--AGAAUCCAUGCAAAACAAGAAGAGCAGAGCUGA (((.......((((...(((((((((......))))))))).-.(---((....))))))).--...((((((.((..--....))..)))....(.....)..))).))).. ( -16.00, z-score = 0.31, R) >droAna3.scaffold_13266 11291772 92 + 19884421 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAAGAUAUUCAAA-AG---UGCGGGCUCGAGAACAAGGCGAGAAAGAAAGGAGAAAGCAAGCAGAGC----------------- .((..............(((((((((......))))))))).-..---(((...(((......................)))...))).)).....----------------- ( -16.45, z-score = -1.82, R) >dp4.chr3 15487099 96 + 19779522 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUACUCGUAUAUAUUCAAAAGUGCGAGGCAUCCAGCGAAAAAAAGGAAGAACACCACCAGAAA----------------- .((....((((((......)))).)).....((((.(((((((..........)))))))..)))).))........((.......))........----------------- ( -16.10, z-score = -1.70, R) >droPer1.super_4 3603270 96 - 7162766 AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUACUCGUAUAUAUUCAAAAGUGCGAGGCAUCCAGCGAAAAAAAGGAAGAACACCACCAGGAA----------------- .((....((((((......)))).)).....((((.(((((((..........)))))))..)))).)).................((....))..----------------- ( -16.50, z-score = -1.47, R) >consensus AGCAUUAGAAUUUGAAAUUGAAUAUCAUAAAGGAUAUUCAGA_AG___UGCGCGCUCGAAAAA_GAACUGGAAAAAAA__AGAAUCCAUGCAAAAC_________________ .................(((((((((......)))))))))........................................................................ ( -7.30 = -7.49 + 0.19)
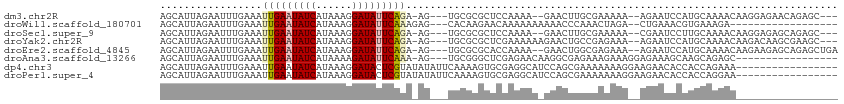
| Location | 17,921,221 – 17,921,323 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.42 |
| Shannon entropy | 0.61895 |
| G+C content | 0.35854 |
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -6.71 |
| Energy contribution | -6.53 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17921221 102 - 21146708 ---GCUCUGUUCUCCUUGUUUUGCAUGGAUUCU--UUUUUCGCAAGUUC--UUUUGGAGCGCGCA---CU-UCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ---...................(((((((....--......((..((((--.....))))..)).---..-..((((((((......)))))))).........))).)))). ( -18.80, z-score = -1.22, R) >droWil1.scaffold_180701 3308199 90 - 3904529 ------------------UCUUUCACGUUUCAG--UCUAGUUUGGGUUUUUUUUUUGUUCUUGUG---CUCUUUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ------------------............((.--...((((((((..........((......)---)...(((((((((......))))))))).))))))))....)).. ( -10.60, z-score = -0.96, R) >droSec1.super_9 1233017 102 - 3197100 ---GCUCUGCUCUCCUUGUUUUGCAAGGAUUCG--UUUUUCGCAAGUUC--UUUUGGAGCGCGCA---CU-UCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ---((..(((((((((((.....))))))....--.......((((...--.))))))))).)).---..-..((((((((......)))))))).................. ( -23.60, z-score = -2.21, R) >droYak2.chr2R 9880771 104 + 21139217 ---GCUUCGCUUGUCUUGUUUUGCAUGGAUUCU--UUUCUCGGCAGUUCUUUUUUCGAGCGCGCA---CU-UCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ---((..((((((....(..((((..(((....--..)))..))))..)......)))))).)).---..-..((((((((......)))))))).................. ( -23.10, z-score = -2.40, R) >droEre2.scaffold_4845 12056500 105 - 22589142 UCAGCUCUGCUCUUCUUGUUUUGCAUGGAUUCU--UUUCUCGCCAGUUC--UUUUGGUGCGCGCA---CU-UCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ......................(((((((....--.....((((((...--..))))))......---..-..((((((((......)))))))).........))).)))). ( -18.20, z-score = -0.78, R) >droAna3.scaffold_13266 11291772 92 - 19884421 -----------------GCUCUGCUUGCUUUCUCCUUUCUUUCUCGCCUUGUUCUCGAGCCCGCA---CU-UUUGAAUAUCUUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU -----------------((..(((..((((..........................))))..)))---..-.(((((((((......)))))))))..............)). ( -13.87, z-score = -2.38, R) >dp4.chr3 15487099 96 - 19779522 -----------------UUUCUGGUGGUGUUCUUCCUUUUUUUCGCUGGAUGCCUCGCACUUUUGAAUAUAUACGAGUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU -----------------......(.(((((((...............))))))).)(((..((((((.......(((((((......)))))))...))))))......))). ( -16.06, z-score = -1.10, R) >droPer1.super_4 3603270 96 + 7162766 -----------------UUCCUGGUGGUGUUCUUCCUUUUUUUCGCUGGAUGCCUCGCACUUUUGAAUAUAUACGAGUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU -----------------......(.(((((((...............))))))).)(((..((((((.......(((((((......)))))))...))))))......))). ( -16.06, z-score = -1.02, R) >consensus _________________GUUUUGCAUGGAUUCU__UUUUUCGCCAGUUC_UUUUUCGAGCGCGCA___CU_UCUGAAUAUCCUUUAUGAUAUUCAAUUUCAAAUUCUAAUGCU ..........................................................................(((((((......)))))))................... ( -6.71 = -6.53 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:22 2011