| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,917,536 – 17,917,647 |
| Length | 111 |
| Max. P | 0.994747 |

| Location | 17,917,536 – 17,917,647 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 67.46 |
| Shannon entropy | 0.62042 |
| G+C content | 0.42753 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -13.17 |
| Energy contribution | -12.49 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17917536 111 - 21146708 AUCCCCAAUCCA--ACUCCUCCCCUCCCACCCACCAACAUGCACAGUCACAAUUUAACGUAUGCGUGAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC ............--...............................(..((((((...((((((((..((.-((.(.......).)).))..))))))))...))))))..)... ( -19.40, z-score = -2.40, R) >droYak2.chr2R 9876920 101 + 21139217 AUCCCCAAU-----AAACCUUCCUCCUGA-------ACAUACACAGUUACAAUUUAACGUAUGCGUGAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC .........-----...........(((.-------.......))).(((((((...((((((((..((.-((.(.......).)).))..))))))))...)))))))..... ( -20.50, z-score = -2.71, R) >droSec1.super_9 1229360 107 - 3197100 AUCCCCAAUCCA--CACACCCCCCCUCCACCC----ACAUGCACAGUCACAAUUUAACGUAUGCGUGAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUAUCACC ............--..................----.........(..((((((...((((((((..((.-((.(.......).)).))..))))))))...))))))..)... ( -19.20, z-score = -2.00, R) >droSim1.chr2R 16559134 109 - 19596830 AUCCCCAAUCCACCAACCCCUCCCCUCCACCC----ACAUGCACAGUCACAAUUUAACGUAUGCGUGAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC ................................----.........(..((((((...((((((((..((.-((.(.......).)).))..))))))))...))))))..)... ( -19.40, z-score = -2.43, R) >droWil1.scaffold_180701 591478 101 + 3904529 -------------CUCACCCGUCACGUCAGCCUUUCCUUUUUGCCAUGGCAUUCCAACGUAUGCGUAAUAUUUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUGCCACU -------------.................................((((((..(..((((((((((((.((((........)))).))))))))))))....)..)))))).. ( -24.90, z-score = -1.73, R) >droVir3.scaffold_12875 11541729 89 - 20611582 -----------------ACUUUUACCCCC-------AACCCUGGAUAAAGUGUUUAAUGUAUAUGUAAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACUGUUGUCCCACU -----------------............-------......((((((((((((((((((.......)))-))))....(((((.......)))))..))))).)))))).... ( -15.90, z-score = -1.43, R) >droAna3.scaffold_13266 11288833 84 - 19884421 --------------------CUCAUCCUG-------CCCUCCUCACUCAC--UUUAACGUAUGCGUGAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACU --------------------.........-------........((..((--(....((((((((..((.-((.(.......).)).))..))))))))...))).))...... ( -15.00, z-score = -1.20, R) >droMoj3.scaffold_6496 11516809 84 + 26866924 ----------------------CACCCGC------AAACUGUGUGUAGAGUAUUUAAUGUAUGUGUAGUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACUGUUGUGCCGC- ----------------------.((.(((------.....))).))............(((((((((((.-((.(.......).)).)))))))))))((((....))))...- ( -19.30, z-score = 0.04, R) >droGri2.scaffold_15245 11643386 90 + 18325388 -----------------ACUCACAUCCGC------AAACUAUAUGUGUACUGUUUAAUGUAUGUGUAAUA-UUGAAACAAUGUGAAAAUUGCGCAUACGCACUGUUGUGACACU -----------------..(((((...((------((((..((....))..))))...(((((((((((.-((.(.......).)).))))))))))))).....))))).... ( -18.10, z-score = 0.06, R) >consensus _________________CCUCCCACCCCA_______ACAUGCACAGUCACAAUUUAACGUAUGCGUGAUA_UUGAAACAAUGUGAAAAUUGCGCAUACGCACAGUUGUACCACC .........................................................((((((((((((..................))))))))))))............... (-13.17 = -12.49 + -0.68)

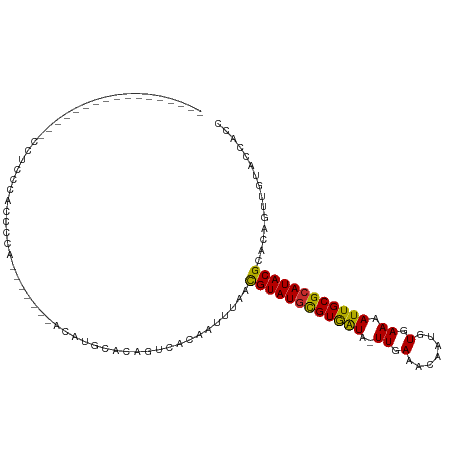
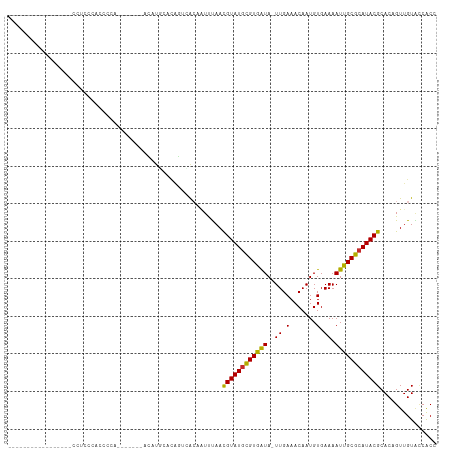
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:19 2011