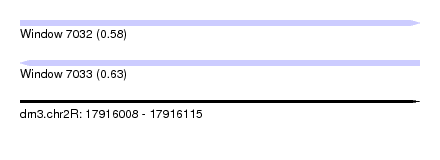
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,916,008 – 17,916,115 |
| Length | 107 |
| Max. P | 0.626592 |

| Location | 17,916,008 – 17,916,115 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.37689 |
| G+C content | 0.35739 |
| Mean single sequence MFE | -20.58 |
| Consensus MFE | -10.39 |
| Energy contribution | -10.94 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
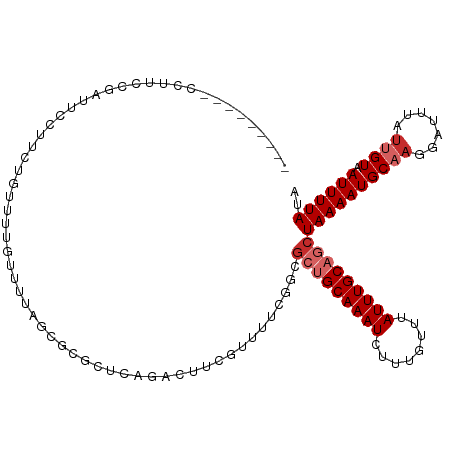
>dm3.chr2R 17916008 107 + 21146708 UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG--------- .................(((((...((((((((((((((........)))))))(((((((........)).).)))))))))))(......))))))..(....).--------- ( -22.90, z-score = -1.94, R) >droSim1.chr2R 16557632 107 + 19596830 UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG--------- .................(((((...((((((((((((((........)))))))(((((((........)).).)))))))))))(......))))))..(....).--------- ( -22.90, z-score = -1.94, R) >droSec1.super_9 1227956 107 + 3197100 UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG--------- .................(((((...((((((((((((((........)))))))(((((((........)).).)))))))))))(......))))))..(....).--------- ( -22.90, z-score = -1.94, R) >droYak2.chr2R 9875410 107 - 21139217 UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG--------- .................(((((...((((((((((((((........)))))))(((((((........)))..)))))))))))(......))))))..(....).--------- ( -23.60, z-score = -2.43, R) >droEre2.scaffold_4845 12051159 107 + 22589142 UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCAGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG--------- .................(((((...((((((((((((((........)))))))(((((((........)))..)))))))))))(......))))))..(....).--------- ( -23.60, z-score = -2.43, R) >droAna3.scaffold_13266 11287608 83 + 19884421 UAUAAAAUUACUAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGC--AGCCGAAA----------------------CAAAGCAGAAGGAGGCAGAAUG--------- .................(((((((.(((..(((((((((........))))))--)))..)))----------------------....)))..)))).........--------- ( -19.80, z-score = -3.27, R) >droPer1.super_4 3598084 116 - 7162766 UAUAAAAUUACAAUAAAUCGUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAACACUGAGCGCGCUAAAACAAAGCAGAAGGAACAGGGAUAUAGGAUAUA ...................(((...((((((((((((((........)))))))((((((...........)).)))))))))))...)))......................... ( -21.00, z-score = -1.10, R) >dp4.chr3 15481988 96 + 19779522 UAUAAAAUUACAAUAAAUCGUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGA--------------------AAGCAGAAGGAACAGGGAUAUAGGAUAUA ................(((.(((.......(((((((((........))))))))).((.....(..--------------------..).....))..))).))).......... ( -19.70, z-score = -2.96, R) >droWil1.scaffold_180701 3314777 81 + 3904529 UAUAAAAUUACAAUAAAUCCAAGCAUUUUAGCUGCAAAUAAACAAAGAUUUGC--CAAC-AAA----------------------CAAAACA-AGUGAGGCGGCAGG--------- ..................((.(((......))).................(((--(..(-..(----------------------(......-.))..)..))))))--------- ( -8.80, z-score = 0.28, R) >consensus UAUAAAAUUACAAUAAAUCCUUGCAUUUUAGCUGCAAAUAAACAAAGAUUUGCAGCGCCGAAAACGAAGUCUGAGCGCGCUAAAACAAAACAGAAGGAAUCGGAAGG_________ ..............................(((((((((........)))))))))............................................................ (-10.39 = -10.94 + 0.56)



| Location | 17,916,008 – 17,916,115 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Shannon entropy | 0.37689 |
| G+C content | 0.35739 |
| Mean single sequence MFE | -22.11 |
| Consensus MFE | -11.26 |
| Energy contribution | -12.14 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17916008 107 - 21146708 ---------CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ---------......((.((((..(((...(((((((((((((.(((......))).)))))(((((((........)))))))))))))))))))))).)).............. ( -26.30, z-score = -2.41, R) >droSim1.chr2R 16557632 107 - 19596830 ---------CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ---------......((.((((..(((...(((((((((((((.(((......))).)))))(((((((........)))))))))))))))))))))).)).............. ( -26.30, z-score = -2.41, R) >droSec1.super_9 1227956 107 - 3197100 ---------CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ---------......((.((((..(((...(((((((((((((.(((......))).)))))(((((((........)))))))))))))))))))))).)).............. ( -26.30, z-score = -2.41, R) >droYak2.chr2R 9875410 107 + 21139217 ---------CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ---------......((.((((..(((...((((((((((((..((((...))))...))))(((((((........)))))))))))))))))))))).)).............. ( -25.30, z-score = -2.61, R) >droEre2.scaffold_4845 12051159 107 - 22589142 ---------CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCUGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ---------......((.((((..(((...((((((((((((..((((...))))...))))(((((((........)))))))))))))))))))))).)).............. ( -25.30, z-score = -2.61, R) >droAna3.scaffold_13266 11287608 83 - 19884421 ---------CAUUCUGCCUCCUUCUGCUUUG----------------------UUUCGGCU--GCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUAGUAAUUUUAUA ---------.........((((..(((...(----------------------(((..(((--((((((........)))))))))..)))))))))))................. ( -20.70, z-score = -2.89, R) >droPer1.super_4 3598084 116 + 7162766 UAUAUCCUAUAUCCCUGUUCCUUCUGCUUUGUUUUAGCGCGCUCAGUGUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAACGAUUUAUUGUAAUUUUAUA ........................(((...(((((((((((((.((........)).)))))(((((((........))))))))))))))))))((((....))))......... ( -23.00, z-score = -1.64, R) >dp4.chr3 15481988 96 - 19779522 UAUAUCCUAUAUCCCUGUUCCUUCUGCUU--------------------UCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAACGAUUUAUUGUAAUUUUAUA .........................(((.--------------------........)))(((((((((........)))))))))((((((((((.......)))).)))))).. ( -15.00, z-score = -1.42, R) >droWil1.scaffold_180701 3314777 81 - 3904529 ---------CCUGCCGCCUCACU-UGUUUUG----------------------UUU-GUUG--GCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCUUGGAUUUAUUGUAAUUUUAUA ---------...((((.(..((.-......)----------------------)..-).))--))((((((.(((......)))((......))..)))))).............. ( -10.80, z-score = 0.56, R) >consensus _________CCUUCCGAUUCCUUCUGUUUUGUUUUAGCGCGCUCAGACUUCGUUUUCGGCGCUGCAAAUCUUUGUUUAUUUGCAGCUAAAAUGCAAGGAUUUAUUGUAAUUUUAUA ............................................................(((((((((........)))))))))((((((((((.......)))).)))))).. (-11.26 = -12.14 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:18 2011