
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,891,060 – 17,891,154 |
| Length | 94 |
| Max. P | 0.856269 |

| Location | 17,891,060 – 17,891,154 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.48108 |
| G+C content | 0.32299 |
| Mean single sequence MFE | -18.97 |
| Consensus MFE | -11.14 |
| Energy contribution | -10.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856269 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
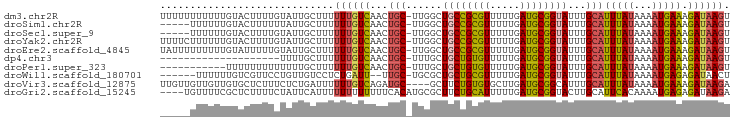
>dm3.chr2R 17891060 94 - 21146708 UUUUUUUUUUUGUACUUUUGUAUUGCUUUUUUGUCAACUGC-UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU ((((((.(((((((....((((..........(((((....-)))))(((((((((...)))))))))..))))..))))))).))))))..... ( -20.90, z-score = -2.00, R) >droSim1.chr2R 16532020 89 - 19596830 -----UUUUUUGUACUUUUUUAUUGCUUUUUUGUCAACUGC-UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU -----...(((((.((((((((((((......(((((....-)))))(((((((((...)))))))))...)))...))))...)))))))))). ( -18.50, z-score = -1.28, R) >droSec1.super_9 1203044 89 - 3197100 -----UUUUUUGUACUUUUGUAUUGCUUUUUUGUCAACUGC-UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU -----(((((..(...((((((.(((......(((((....-)))))(((((((((...)))))))))...)))..)))))))..)))))..... ( -20.50, z-score = -1.80, R) >droYak2.chr2R 9849244 94 + 21139217 UUUUCUUUUUUGUACUUUUGUAUUGCUUUUUUGUCAACUGC-UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU ...((((((......(((((((.(((......(((((....-)))))(((((((((...)))))))))...)))..))))))).))))))..... ( -21.80, z-score = -2.35, R) >droEre2.scaffold_4845 12025991 94 - 22589142 UAUUUUUUUUUGUAUUUUUGUAUUGCUUUUUUGUCAACUGC-UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU ((((((((.......(((((((.(((......(((((....-)))))(((((((((...)))))))))...)))..))))))).))))))))... ( -21.70, z-score = -1.97, R) >dp4.chr3 15454923 74 - 19779522 --------------------UUUUGCUUUUUUGUCAACUGC-UUUGCUGCUGUGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU --------------------.........((((((...(((-..((((((............))))))...)))(((((...))))).)))))). ( -12.40, z-score = 0.03, R) >droPer1.super_323 483 83 - 21222 -----------UUUUUUUUUUUUUGCUUUUUUGUCAACUGC-UUUGCUGCUGUGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU -----------.......((((((....((((((.((.(((-..((((((............))))))...))).)))))))).))))))..... ( -12.70, z-score = 0.23, R) >droWil1.scaffold_180701 3268546 86 - 3904529 ------UUUUUUGUCGUUCCUGUUGUCCUCUGAUU--UUGC-UGCGCUGCUGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAGAGAUAACU ------...............((((((.(((.(((--(((.-((((.(((((((((...)))))))))..))))....)))))).))))))))). ( -23.90, z-score = -3.46, R) >droVir3.scaffold_12875 11509968 91 - 20611582 UUGUUGUUGUUGUGCUCUUUCUCUGAUUUUUUGUCAGAUGC----GCUUCUGUGUGCUUGAUGCGGCAUUUGCAUUUAUAAAAUGAAAGAUAAGA ((((...(((.(((((...((((((((.....)))))).((----((......))))..))...)))))..)))...)))).............. ( -19.70, z-score = -0.66, R) >droGri2.scaffold_15245 11608760 91 + 18325388 ----UGUUUUCGCUCUUUUCUAUUCAUUUUUUUUUUUUCACAUGCGCUUCUGCAUUUUUGAUGCGGUACUUGCAUUCACAAAAUGAGAGAUAAGA ----.........((((.(((.((((((((...........(((((...(((((((...)))))))....)))))....))))))))))).)))) ( -17.56, z-score = -1.32, R) >consensus _____UUUUUUGUACUUUUGUAUUGCUUUUUUGUCAACUGC_UUGGCUGCCGCGUUUUUGAUGCGGUAUUUGCAUUUAUAAAAUGAAAGAUAAGU .............................((((((..........((((((((((.....))))))))...)).(((((...))))).)))))). (-11.14 = -10.70 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:15 2011