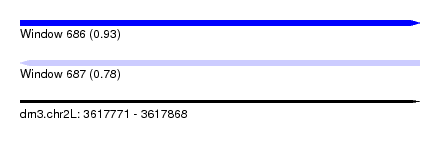
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,617,771 – 3,617,868 |
| Length | 97 |
| Max. P | 0.934342 |

| Location | 3,617,771 – 3,617,868 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 60.42 |
| Shannon entropy | 0.77310 |
| G+C content | 0.57531 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -9.39 |
| Energy contribution | -9.88 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
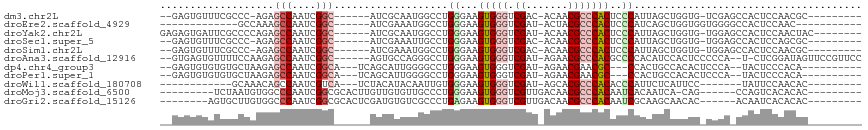
>dm3.chr2L 3617771 97 + 23011544 --GAGUGUUUCGCCC-AGAGCCAAUCGGC------AUCGCAAUGGCCUGGGAAGUGGGUCGAC-ACAACGCCCACUCCCAUUAGCUGGUG-UCGAGCCACUCCAACGC--------- --(((((.(((((((-((.((((....((------...))..)))).(((((.(((((.((..-....))))))))))))....)))).)-.)))).)))))......--------- ( -40.30, z-score = -2.83, R) >droEre2.scaffold_4929 3656232 86 + 26641161 -------------GCCAAAGCCAAUCGGC------AUCGAAAUGGCCUGGGAAGUGGGUCGAU-ACUACGCCCACUCCCAUCAGCUGGUGGUGGGGCCACUCCAAC----------- -------------......(((....)))------.......((((((((((.(((((.((..-....)))))))))))..((.(....).)))))))).......----------- ( -33.50, z-score = -1.40, R) >droYak2.chr2L 3611111 101 + 22324452 GAGAGUGAUUCGCCCCAGAGCCAAUCGGC------AUCGCAAUGGCCUGGGAAGUGGGUCGAU-ACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAACUAC-------- (.(((((.(((((.((((.((((....((------...))..)))).(((((.(((((.((..-....))))))))))))....)))).)-))))..))))))......-------- ( -42.10, z-score = -3.17, R) >droSec1.super_5 1718733 97 + 5866729 --GAGUGUUUCGCCC-AGAGCCAAUCGGC------AUCGAAAUUGCCUGGGAAGUGGGUCGAC-ACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAGCGC--------- --(((((((((((((-((........(((------(.......))))(((((.(((((.((..-....))))))))))))....)))).)-))))).)))))......--------- ( -43.00, z-score = -3.58, R) >droSim1.chr2L 3576857 97 + 22036055 --GAGUGUUUCGCCC-AGAGCCAAUCGGC------AUCGAAAUGGCCUGGGAAGUGGGUCGAC-ACAACGCCCACUCCCAUUAGCUGGUG-UGGAGCCACUCCAACGC--------- --(((((((((((((-((.((((.(((..------..)))..)))).(((((.(((((.((..-....))))))))))))....)))).)-))))).)))))......--------- ( -43.80, z-score = -3.69, R) >droAna3.scaffold_12916 5670656 105 + 16180835 --GUGAGUGUUUUCCAAGAGCCAAUCGGC------AGUGCCAGGGCCUGGGAAGUGGGUCGAU-AGAACGCCCACGCCCCACAUCCACUCCCCCA--U-CUCGGAUAGUUCCGUUCC --..(((((.......((.(((....(((------...)))..)))))(((..(((((.((..-....)))))))..))).....))))).....--.-..((((....)))).... ( -34.30, z-score = -0.95, R) >dp4.chr4_group3 6569188 96 - 11692001 --GAGUGUGUGUGCUAAGAGCCAAUCGGCA---UCAGCAUUGGGGCCUGGGAAGUGGGUCGAU-AGAACGAACGC---CCACUGCCACACUCCCA--UACUCCCACA---------- --(((((((.(((((..(((((....))).---))))))).((((..(((..(((((((((..-....))...))---))))).)))..))))))--))))).....---------- ( -36.90, z-score = -1.88, R) >droPer1.super_1 3668895 96 - 10282868 --GAGUGUGUGUGCUAAGAGCCAAUCGGCA---UCAGCAUUGGGGCCUGGGAAGUGGGUCGAU-AGAACGAACGC---CCACUGCCACACUCCCA--UACUCCCACA---------- --(((((((.(((((..(((((....))).---))))))).((((..(((..(((((((((..-....))...))---))))).)))..))))))--))))).....---------- ( -36.90, z-score = -1.88, R) >droWil1.scaffold_180708 3018823 85 - 12563649 ------------GCAAACAGCCAAUCGUCA---UCUACAUACAAUUGUGGGAAGUGGGUCGAU-AGCACGCCCACACCCAUUCUCAUUCC-------UAUUCCAACAC--------- ------------((.....)).........---.............(((((..(((((.((..-....))))))).))))).........-------...........--------- ( -16.30, z-score = -1.12, R) >droMoj3.scaffold_6500 14240630 92 - 32352404 ---------UCUAAUGUGGCCCAAUCGGCGCACUUGUUGUGUUGCCCUGGGAAGUGGGUCGUUGACAACGCCCACAAUCACAAUCA-CAG------CCAGUCACACAC--------- ---------.....(((((((.....)))(.(((.(((((((((...(((...(((((.(((.....))))))))..)))))).))-)))------).)))).)))).--------- ( -30.10, z-score = -1.01, R) >droGri2.scaffold_15126 4263571 94 + 8399593 --------AGUGCUUGUGGCCCAAUCGGCGCACUCGAUGUGUCGCCCUGAGAAGUGGGUCGUUGACAACGCCCACAAUCGCAAGCAACAC------ACAAUCACACAC--------- --------..(((((((((.......(((((((.....))).)))).......(((((.(((.....))))))))..)))))))))....------............--------- ( -34.40, z-score = -2.14, R) >consensus __GAGUGUUUCGCCCAAGAGCCAAUCGGC______AUCGUAAUGGCCUGGGAAGUGGGUCGAU_ACAACGCCCACUCCCAUUAGCUGCUG_UCGA_CCACUCCAACAC_________ ...................(((....)))...................((...(((((.((.......)))))))..))...................................... ( -9.39 = -9.88 + 0.49)
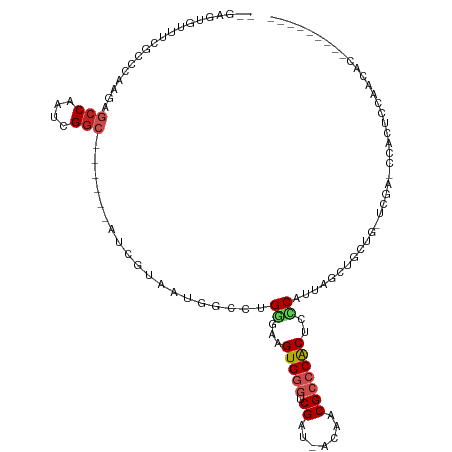
| Location | 3,617,771 – 3,617,868 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 60.42 |
| Shannon entropy | 0.77310 |
| G+C content | 0.57531 |
| Mean single sequence MFE | -38.66 |
| Consensus MFE | -8.83 |
| Energy contribution | -9.65 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3617771 97 - 23011544 ---------GCGUUGGAGUGGCUCGA-CACCAGCUAAUGGGAGUGGGCGUUGU-GUCGACCCACUUCCCAGGCCAUUGCGAU------GCCGAUUGGCUCU-GGGCGAAACACUC-- ---------......(((((..(((.-..((((....((((((((((((....-..)).))))).)))))(((((...((..------..))..)))))))-)).)))..)))))-- ( -43.40, z-score = -2.57, R) >droEre2.scaffold_4929 3656232 86 - 26641161 -----------GUUGGAGUGGCCCCACCACCAGCUGAUGGGAGUGGGCGUAGU-AUCGACCCACUUCCCAGGCCAUUUCGAU------GCCGAUUGGCUUUGGC------------- -----------((((((((((((.((.(....).)).((((((((((((....-..)).))))).)))))))))))))))))------(((((......)))))------------- ( -40.00, z-score = -3.22, R) >droYak2.chr2L 3611111 101 - 22324452 --------GUAGUUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGU-AUCGACCCACUUCCCAGGCCAUUGCGAU------GCCGAUUGGCUCUGGGGCGAAUCACUCUC --------.(((((((.(((....))-).)))))))..(((((((((.((((.-..)))))).(.((((((((((...((..------..))..)))).)))))).)...))))))) ( -44.70, z-score = -2.79, R) >droSec1.super_5 1718733 97 - 5866729 ---------GCGCUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGU-GUCGACCCACUUCCCAGGCAAUUUCGAU------GCCGAUUGGCUCU-GGGCGAAACACUC-- ---------(((((((.(((....))-).)))))...((((((((((((....-..)).))))).))))).))..(((((..------(((....)))...-...))))).....-- ( -41.40, z-score = -2.37, R) >droSim1.chr2L 3576857 97 - 22036055 ---------GCGUUGGAGUGGCUCCA-CACCAGCUAAUGGGAGUGGGCGUUGU-GUCGACCCACUUCCCAGGCCAUUUCGAU------GCCGAUUGGCUCU-GGGCGAAACACUC-- ---------(((((((.(((....))-).)))))....(((((((((((....-..)).)))))))))(((((((..(((..------..))).)))).))-).)).........-- ( -43.10, z-score = -2.78, R) >droAna3.scaffold_12916 5670656 105 - 16180835 GGAACGGAACUAUCCGAG-A--UGGGGGAGUGGAUGUGGGGCGUGGGCGUUCU-AUCGACCCACUUCCCAGGCCCUGGCACU------GCCGAUUGGCUCUUGGAAAACACUCAC-- ............((((((-(--..(((((((((..((((((((....))))))-))....)))))))))..(((.((((...------))))...))))))))))..........-- ( -41.10, z-score = -1.24, R) >dp4.chr4_group3 6569188 96 + 11692001 ----------UGUGGGAGUA--UGGGAGUGUGGCAGUGG---GCGUUCGUUCU-AUCGACCCACUUCCCAGGCCCCAAUGCUGA---UGCCGAUUGGCUCUUAGCACACACACUC-- ----------((((......--((((.((.(((.(((((---(...(((....-..)))))))))..))).)))))).((((((---.(((....)))..)))))))))).....-- ( -38.10, z-score = -1.69, R) >droPer1.super_1 3668895 96 + 10282868 ----------UGUGGGAGUA--UGGGAGUGUGGCAGUGG---GCGUUCGUUCU-AUCGACCCACUUCCCAGGCCCCAAUGCUGA---UGCCGAUUGGCUCUUAGCACACACACUC-- ----------((((......--((((.((.(((.(((((---(...(((....-..)))))))))..))).)))))).((((((---.(((....)))..)))))))))).....-- ( -38.10, z-score = -1.69, R) >droWil1.scaffold_180708 3018823 85 + 12563649 ---------GUGUUGGAAUA-------GGAAUGAGAAUGGGUGUGGGCGUGCU-AUCGACCCACUUCCCACAAUUGUAUGUAGA---UGACGAUUGGCUGUUUGC------------ ---------......(((((-------(.........((((.(((((((....-..)).)))))..))))(((((((.......---..))))))).))))))..------------ ( -26.00, z-score = -1.80, R) >droMoj3.scaffold_6500 14240630 92 + 32352404 ---------GUGUGUGACUGG------CUG-UGAUUGUGAUUGUGGGCGUUGUCAACGACCCACUUCCCAGGGCAACACAACAAGUGCGCCGAUUGGGCCACAUUAGA--------- ---------.(((((.....(------((.-((...(.((..((((((((.....))).))))).))))).))).)))))....(((.(((.....))))))......--------- ( -29.50, z-score = 0.24, R) >droGri2.scaffold_15126 4263571 94 - 8399593 ---------GUGUGUGAUUGU------GUGUUGCUUGCGAUUGUGGGCGUUGUCAACGACCCACUUCUCAGGGCGACACAUCGAGUGCGCCGAUUGGGCCACAAGCACU-------- ---------..((((..((((------((((((((((.((..((((((((.....))).))))).)).)).))))))))).)))(((.(((.....))))))..)))).-------- ( -39.90, z-score = -2.15, R) >consensus _________GUGUUGGAGUGG_UCCA_CACCAGCUAAUGGGAGUGGGCGUUGU_AUCGACCCACUUCCCAGGCCAUUACGAU______GCCGAUUGGCUCUUGGGCGAAACACUC__ ..............(((((...................((..(((((((.......)).)))))...)).((.................)).....)))))................ ( -8.83 = -9.65 + 0.82)

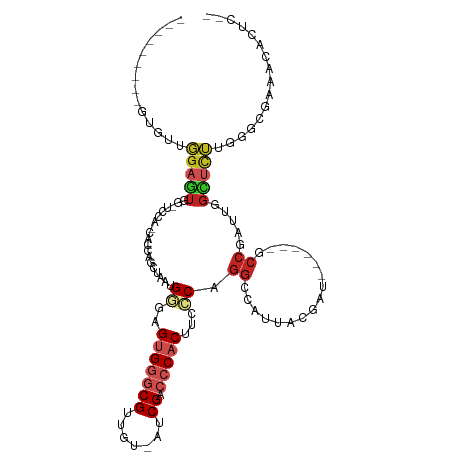
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:13 2011