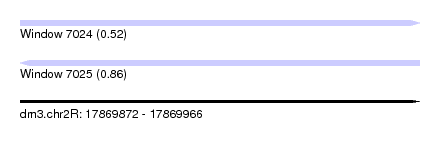
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,869,872 – 17,869,966 |
| Length | 94 |
| Max. P | 0.861369 |

| Location | 17,869,872 – 17,869,966 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Shannon entropy | 0.38537 |
| G+C content | 0.54247 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -19.60 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
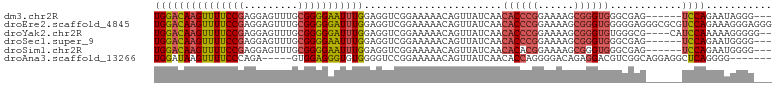
>dm3.chr2R 17869872 94 + 21146708 UGGACAAGUUUUCCGAGGAGUUUGCGGGGAAUUUGGAGGUCGGAAAAACAGUUAUCAACACCCGGAAAAGCGGGUGGGCGAG------UCCAGAAUAGGG--- ((((((((((..(((.........)))..)))))....((((......).........((((((......)))))))))..)------))))........--- ( -28.90, z-score = -2.00, R) >droEre2.scaffold_4845 12004392 103 + 22589142 UGGACAAGUUUUCCGAGGAGUUUGCGGGGGAUUUGGAGGUCGGAAAAACAGUUAUCAACACCCGGAAAAGCGGGUGGGGGAGGGCGCGUCCAGAAAGGGAGGG ((((((((((..(((.........)))..)))))................((..((..((((((......))))))...))..))..)))))........... ( -32.00, z-score = -1.82, R) >droYak2.chr2R 9826824 97 - 21139217 UGGACAAGUUUUCCGAGGAGUUUGCGGGGGAUUUGGAGGUCGGAAAAACAGUUAUCAACACCCGGAAAAGCGGGUGUGGGCG----CAUCCAAAAAGGGGG-- ((((((((((..(((.........)))..))))))...............(((....(((((((......))))))).))).----..)))).........-- ( -29.90, z-score = -1.60, R) >droSec1.super_9 1181931 94 + 3197100 UGGACAAGUUUUCCGAGGAGUUUGCGGGGAAUUUGGAGGUCGGAAAAACAGUUAUCAACACCCGGAAAAGCGGGUGGGCGAG------UCCAGAAUGGGG--- ((((((((((..(((.........)))..)))))....((((......).........((((((......)))))))))..)------))))........--- ( -28.90, z-score = -1.62, R) >droSim1.chr2R 16510224 94 + 19596830 UGGACAAGUUUUCCGAGGAGUUUGCGGGGAAUUUGGAGGUCGGAAAAACAGUUAUCAACACACGGAAAAGCGGGUGGGCGAG------UCCAGAAUGGGG--- ((((((((((..(((.........)))..)))))....((((......).........(((.((......)).))))))..)------))))........--- ( -21.90, z-score = -0.28, R) >droAna3.scaffold_13266 11244669 91 + 19884421 UGGAUAAGUUUUCCCAGA-----GUGGAGGGUGUGGGGUCCGGAAAAACAGUUAUCAACACCAGGGGACAGAGGACGUCGGCAGGAGGCUCAGGGG------- ............(((.((-----((.....((.(((.((((((.................)).(....)...)))).))))).....)))).))).------- ( -21.13, z-score = 1.00, R) >consensus UGGACAAGUUUUCCGAGGAGUUUGCGGGGAAUUUGGAGGUCGGAAAAACAGUUAUCAACACCCGGAAAAGCGGGUGGGCGAG______UCCAGAAUGGGG___ (((((((((((((((.........))))))))))).......................((((((......))))))............))))........... (-19.60 = -20.13 + 0.53)


| Location | 17,869,872 – 17,869,966 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.52 |
| Shannon entropy | 0.38537 |
| G+C content | 0.54247 |
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -12.83 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17869872 94 - 21146708 ---CCCUAUUCUGGA------CUCGCCCACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCA ---.........(((------((((..((((((......)))))).))).......((((((((.......................))))))))...)))). ( -22.00, z-score = -2.60, R) >droEre2.scaffold_4845 12004392 103 - 22589142 CCCUCCCUUUCUGGACGCGCCCUCCCCCACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUCCCCCGCAAACUCCUCGGAAAACUUGUCCA ............((((...........((((((......))))))...........((((((((.......................))))))))...)))). ( -22.40, z-score = -2.45, R) >droYak2.chr2R 9826824 97 + 21139217 --CCCCCUUUUUGGAUG----CGCCCACACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUCCCCCGCAAACUCCUCGGAAAACUUGUCCA --.........(((((.----...(.(((((((......))))))).)........((((((((.......................))))))))...))))) ( -22.00, z-score = -1.90, R) >droSec1.super_9 1181931 94 - 3197100 ---CCCCAUUCUGGA------CUCGCCCACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCA ---.........(((------((((..((((((......)))))).))).......((((((((.......................))))))))...)))). ( -22.00, z-score = -2.38, R) >droSim1.chr2R 16510224 94 - 19596830 ---CCCCAUUCUGGA------CUCGCCCACCCGCUUUUCCGUGUGUUGAUAACUGUUUUUCCGACCUCCAAAUUCCCCGCAAACUCCUCGGAAAACUUGUCCA ---.........(((------((((..(((.((......)).))).))).......((((((((.......................))))))))...)))). ( -14.90, z-score = -0.97, R) >droAna3.scaffold_13266 11244669 91 - 19884421 -------CCCCUGAGCCUCCUGCCGACGUCCUCUGUCCCCUGGUGUUGAUAACUGUUUUUCCGGACCCCACACCCUCCAC-----UCUGGGAAAACUUAUCCA -------......(((..((....((((.....))))....)).)))(((((..(((((((((((...............-----)))))))))))))))).. ( -18.26, z-score = -0.64, R) >consensus ___CCCCAUUCUGGA______CUCGCCCACCCGCUUUUCCGGGUGUUGAUAACUGUUUUUCCGACCUCCAAAUCCCCCGCAAACUCCUCGGAAAACUUGUCCA ...........((((............((((((......))))))...........((((((((.......................))))))))....)))) (-12.83 = -13.72 + 0.89)

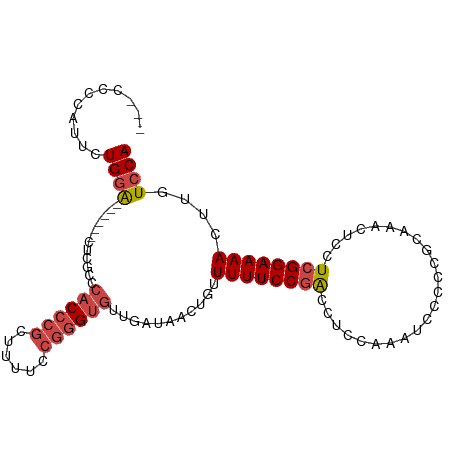
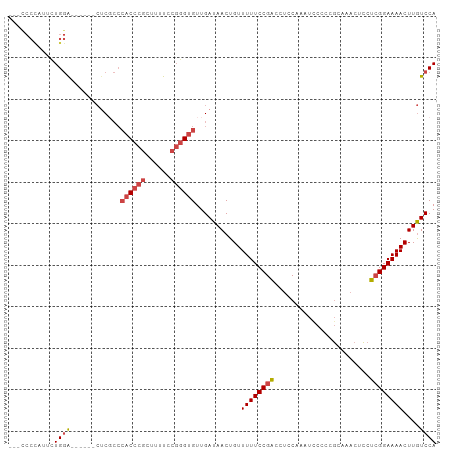
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:11 2011