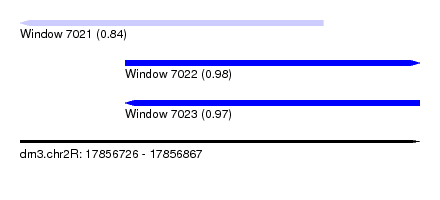
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,856,726 – 17,856,867 |
| Length | 141 |
| Max. P | 0.980384 |

| Location | 17,856,726 – 17,856,833 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.74 |
| Shannon entropy | 0.16042 |
| G+C content | 0.43340 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.59 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17856726 107 - 21146708 GUCAACCUGUCCGUAAUCUGGUCUC-CCCAGCAAAUUACUUCCCGCUUUUCAAUUACGCAACAGGUUGUCUAUUUCAACCUUGACUUUUGACCAUCCUACGG---AAAAUU .........((((((...(((((..-.....(((..........((...........))....(((((.......))))))))......)))))...)))))---)..... ( -21.72, z-score = -2.71, R) >droSim1.chr2R 16493205 105 - 19596830 GUCAACCUGU-CGUAAUCUGGUCUCGCCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCUAUUUCAA-CUUGACUUUUGACCA-CCUACGG---AAAAUU (.((((((((-.(((((((((......))))...))))).....((...........)).)))))))).)....((((-........))))((.-.....))---...... ( -19.80, z-score = -1.43, R) >droSec1.super_9 1169205 108 - 3197100 GUCAACCUGUCCGUAAUCUGGUCUCGCCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCUAUUUCAACCUUGACUUUUGACCACCCUACGG---AAAAUU .........((((((...(((((......(((............)))...............((((((.......))))))........)))))...)))))---)..... ( -21.60, z-score = -2.06, R) >droYak2.chr2R 9812938 111 + 21139217 GUCAACCUGUCUGUAAUCUUGUCUCCGCCAGCAAAUGACUUCCACCUUUUCAAUUACGCAACAGGUUGUCUAUUUCAACCUUGACUUUUGACCACCCCACCACCCCAAAUU (((((...(((.(((((.((((........)))).(((...........)))))))).....((((((.......)))))).)))..)))))................... ( -15.10, z-score = -1.27, R) >droEre2.scaffold_4845 11988110 111 - 22589142 GUCAACCUGUCUGUAAUCUUGUCUCCCCCAGCAAAUUACUUCCACCUAUUCAAUUACGCAACAGGUUGUCUAUUUCAACCUUGACUUUUGACCACCCUGCCACCCUAAAUU (((((...(((.(((((.((((........))))))))).......................((((((.......)))))).)))..)))))................... ( -17.30, z-score = -2.93, R) >consensus GUCAACCUGUCCGUAAUCUGGUCUCCCCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCUAUUUCAACCUUGACUUUUGACCACCCUACGG___AAAAUU ((((((((((.((((((((((......)))).....................))))))..)))))))(((............)))....)))................... (-15.43 = -15.59 + 0.16)

| Location | 17,856,763 – 17,856,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Shannon entropy | 0.33708 |
| G+C content | 0.39942 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
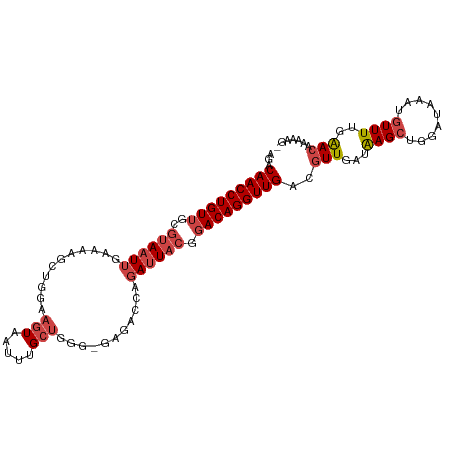
>dm3.chr2R 17856763 104 + 21146708 AGACAACCUGUUGCGUAAUUGAAAAGCGGGAAGUAAUUUGCUGGG-GAGACCAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAAAAG- ...(((((((((.(((((((....((((((......)))))).((-....)).))))))))))))))))..(((.(.((((.........))))).)))......- ( -30.40, z-score = -4.04, R) >droSim1.chr2R 16493240 102 + 19596830 AGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGGCGAGACCAGAUUACG-ACAGGUUGACGUUGA-AAGCUGGAUAA-UGUUUUGAACAAAAAG- ...((((((((..(((((((.....(((...(((.....))).))).......)))))))-))))))))..(((.(-((((.......-.))))).)))......- ( -29.00, z-score = -3.34, R) >droSec1.super_9 1169242 105 + 3197100 AGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGGCGAGACCAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUAAACAAAAAG- ...(((((((((.(((((((.....(((...(((.....))).))).......))))))))))))))))..(((...((((.........))))..)))......- ( -27.30, z-score = -2.71, R) >droYak2.chr2R 9812978 105 - 21139217 AGACAACCUGUUGCGUAAUUGAAAAGGUGGAAGUCAUUUGCUGGCGGAGACAAGAUUACAGACAGGUUGACGUUGAUAAGCGGGAUAAAUGUUUUGAACAAAAAG- ...(((((((((..((((((............((((.....))))(....)..)))))).))))))))).((((....)))).......................- ( -26.70, z-score = -2.45, R) >droEre2.scaffold_4845 11988150 106 + 22589142 AGACAACCUGUUGCGUAAUUGAAUAGGUGGAAGUAAUUUGCUGGGGGAGACAAGAUUACAGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAAAAAG ...(((((((((..((((((.....((..((.....))..))...(....)..)))))).)))))))))..(((.(.((((.........))))).)))....... ( -25.30, z-score = -2.77, R) >droAna3.scaffold_13266 11227997 88 + 19884421 -GACAACCUGUUGAAUAUUUGAC-------GAGUAAUUUG------GAAAU---AUUUCGGACAGGUUGACUUUGACGAGAUGCUUAAAUAUUUCCGAUAGCAGG- -.....(((((((..(((((...-------)))))..(((------(((((---((((.((.((..(((.......)))..)))).)))))))))))))))))))- ( -22.10, z-score = -2.92, R) >consensus AGACAACCUGUUGCGUAAUUGAAAAGCUGGAAGUAAUUUGCUGGG_GAGACCAGAUUACGGACAGGUUGACGUUGAUAAGCUGGAUAAAUGUUUUGAACAAAAAG_ ...((((((((...((((((...........(((.....)))...........))))))..))))))))..(((....)))......................... (-14.21 = -14.93 + 0.72)
| Location | 17,856,763 – 17,856,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Shannon entropy | 0.33708 |
| G+C content | 0.39942 |
| Mean single sequence MFE | -16.26 |
| Consensus MFE | -11.19 |
| Energy contribution | -11.38 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
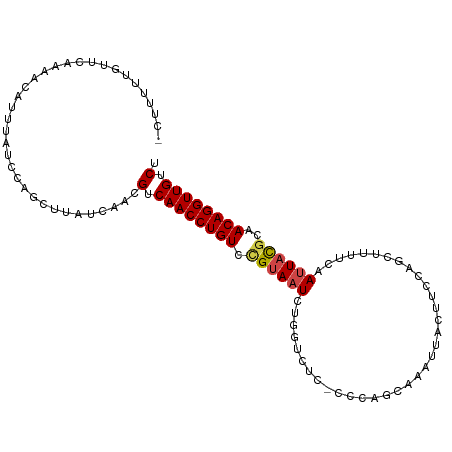
>dm3.chr2R 17856763 104 - 21146708 -CUUUUUGUUCAAAACAUUUAUCCAGCUUAUCAACGUCAACCUGUCCGUAAUCUGGUCUC-CCCAGCAAAUUACUUCCCGCUUUUCAAUUACGCAACAGGUUGUCU -..................................(.((((((((..(((((((((....-.))))...))))).....((...........)).)))))))).). ( -18.80, z-score = -3.56, R) >droSim1.chr2R 16493240 102 - 19596830 -CUUUUUGUUCAAAACA-UUAUCCAGCUU-UCAACGUCAACCUGU-CGUAAUCUGGUCUCGCCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCU -................-...........-.....(.((((((((-.(((((((((......))))...))))).....((...........)).)))))))).). ( -18.90, z-score = -2.86, R) >droSec1.super_9 1169242 105 - 3197100 -CUUUUUGUUUAAAACAUUUAUCCAGCUUAUCAACGUCAACCUGUCCGUAAUCUGGUCUCGCCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCU -..................................(.((((((((..(((((((((......))))...))))).....((...........)).)))))))).). ( -18.90, z-score = -2.80, R) >droYak2.chr2R 9812978 105 + 21139217 -CUUUUUGUUCAAAACAUUUAUCCCGCUUAUCAACGUCAACCUGUCUGUAAUCUUGUCUCCGCCAGCAAAUGACUUCCACCUUUUCAAUUACGCAACAGGUUGUCU -..................................(.((((((((.((((((.((((........)))).(((...........)))))))))..)))))))).). ( -14.80, z-score = -1.45, R) >droEre2.scaffold_4845 11988150 106 - 22589142 CUUUUUUGUUCAAAACAUUUAUCCAGCUUAUCAACGUCAACCUGUCUGUAAUCUUGUCUCCCCCAGCAAAUUACUUCCACCUAUUCAAUUACGCAACAGGUUGUCU ...................................(.((((((((..(((((.((((........))))))))).....................)))))))).). ( -13.40, z-score = -2.12, R) >droAna3.scaffold_13266 11227997 88 - 19884421 -CCUGCUAUCGGAAAUAUUUAAGCAUCUCGUCAAAGUCAACCUGUCCGAAAU---AUUUC------CAAAUUACUC-------GUCAAAUAUUCAACAGGUUGUC- -..((((..............))))..........(.((((((((..(((.(---((((.------..........-------...)))))))).)))))))).)- ( -12.78, z-score = -1.52, R) >consensus _CUUUUUGUUCAAAACAUUUAUCCAGCUUAUCAACGUCAACCUGUCCGUAAUCUGGUCUC_CCCAGCAAAUUACUUCCAGCUUUUCAAUUACGCAACAGGUUGUCU ...................................(.((((((((.((((((...................................))))))..)))))))).). (-11.19 = -11.38 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:10 2011